Defining RNA-seq (gene function) based Tracks¶
tldr 4 "new" tracks
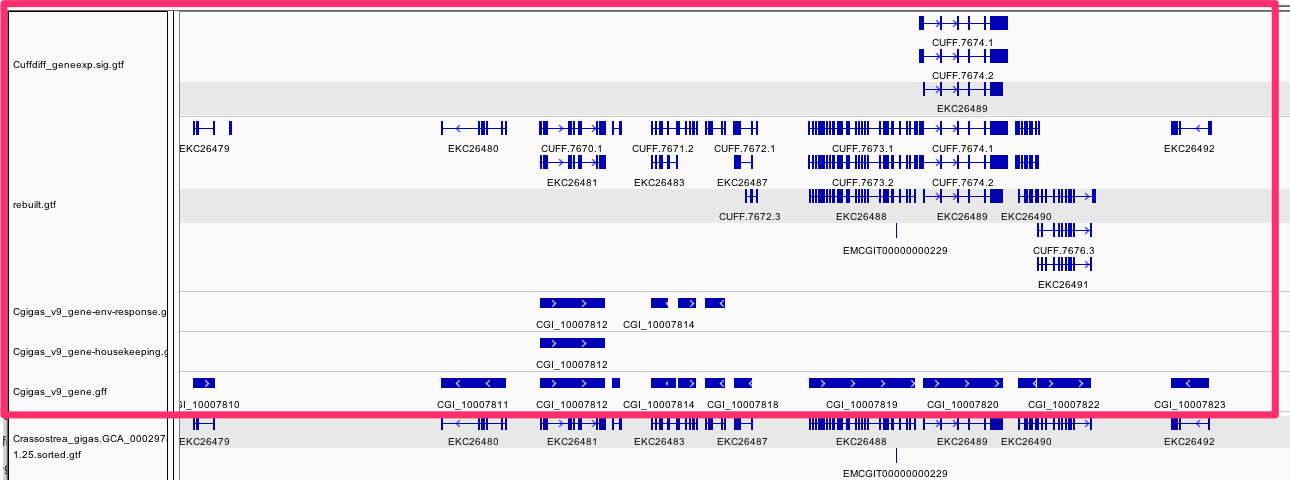 ```
/Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf
/Users/sr320/data-genomic/tentacle/rebuilt.gtf
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff
```
```
/Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf
/Users/sr320/data-genomic/tentacle/rebuilt.gtf
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff
```
Diff Exp Genes¶
In [96]:
#Track with DEGs defined by Cuffdiff
#how derived = {RNA-seq-Gene-ID}
!tail -3 /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf
In [9]:
!wc -l /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf
122038 /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf
New GTF from Cuffdiff¶
In [6]:
#GTF produced from Cuffdiff
#see /Volumes/web/halfshell/BS-heat/Cuffdiff2_heat-b-2014-12-20-22-27-15.4
!head -3 /Users/sr320/data-genomic/tentacle/rebuilt.gtf
C12764 Cufflinks exon 28 201 . . . gene_id XLOC_000001; tss_id "TSS1"; oId "CUFF.1.1"; exon_number "1"; class_code "u"; transcript_id "TCONS_00000001" C12764 Cufflinks CDS 28 201 . . . gene_id XLOC_000001; tss_id "TSS1"; oId "CUFF.1.1"; exon_number "1"; class_code "u"; transcript_id "TCONS_00000001" C12768 Cufflinks exon 4 189 . . . gene_id XLOC_000002; tss_id "TSS2"; oId "CUFF.2.1"; exon_number "1"; class_code "u"; transcript_id "TCONS_00000002"
In [7]:
!wc -l /Users/sr320/data-genomic/tentacle/rebuilt.gtf
1347244 /Users/sr320/data-genomic/tentacle/rebuilt.gtf
GigaDB gene tracks - Isolated Housekeeping and Environment Stress Genes¶
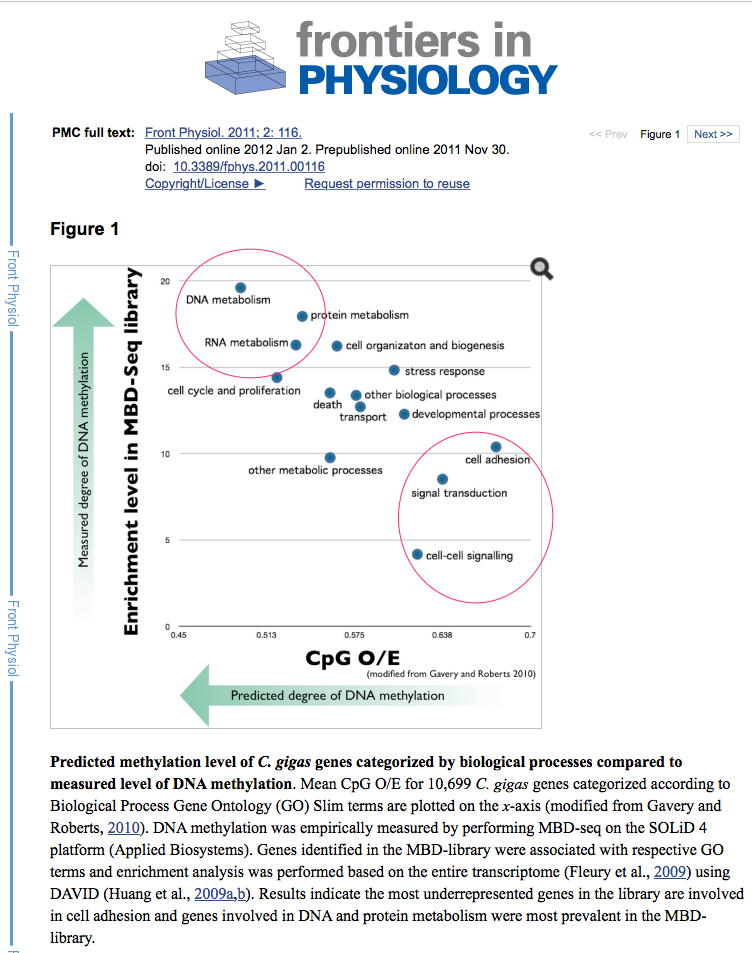
Based on annotation from 10.3389/fphys.2011.00116 (see image above)
In [12]:
!head -3 /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_gene.gff
C16582 GLEAN mRNA 35 385 0.555898 - . ID=CGI_10000001; C17212 GLEAN mRNA 31 363 0.999572 + . ID=CGI_10000002; C17316 GLEAN mRNA 30 257 0.555898 + . ID=CGI_10000003;
In [13]:
!wc -l /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_gene.gff
28027 /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_gene.gff
In [30]:
#adding extra CGI column to join GO info on
!awk -F["\t"] '{print $9}' /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_gene.gff \
| rev | cut -c 2- | rev | sed s/ID=C/C/g > \
/Users/sr320/data-genomic/tentacle/Cgigas_v9_cgi
!head /Users/sr320/data-genomic/tentacle/Cgigas_v9_cgi
CGI_10000001 CGI_10000002 CGI_10000003 CGI_10000004 CGI_10000005 CGI_10000009 CGI_10000010 CGI_10000011 CGI_10000012 CGI_10000013
In [32]:
!wc -l /Users/sr320/data-genomic/tentacle/Cgigas_v9_cgi
28027 /Users/sr320/data-genomic/tentacle/Cgigas_v9_cgi
In [33]:
!paste /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_gene.gff \
/Users/sr320/data-genomic/tentacle/Cgigas_v9_cgi \
> /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID.tab
In [34]:
!head /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID.tab
C16582 GLEAN mRNA 35 385 0.555898 - . ID=CGI_10000001; CGI_10000001 C17212 GLEAN mRNA 31 363 0.999572 + . ID=CGI_10000002; CGI_10000002 C17316 GLEAN mRNA 30 257 0.555898 + . ID=CGI_10000003; CGI_10000003 C17476 GLEAN mRNA 34 257 0.998947 - . ID=CGI_10000004; CGI_10000004 C17998 GLEAN mRNA 196 387 1 - . ID=CGI_10000005; CGI_10000005 C18346 GLEAN mRNA 174 551 1 + . ID=CGI_10000009; CGI_10000009 C18428 GLEAN mRNA 286 546 0.555898 - . ID=CGI_10000010; CGI_10000010 C18964 GLEAN mRNA 203 658 0.999572 - . ID=CGI_10000011; CGI_10000011 C18980 GLEAN mRNA 30 674 0.555898 + . ID=CGI_10000012; CGI_10000012 C19100 GLEAN mRNA 160 681 0.999955 - . ID=CGI_10000013; CGI_10000013
In [35]:
sqls="/Applications/bioinfo/sqlshare-pythonclient/tools/"
In [36]:
!python {sqls}singleupload.py \
-d Cgigas_v9_gene--ID \
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID.tab
processing chunk line 0 to 28027 (0.00476694107056 s elapsed) pushing /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID.tab... parsing 0863C50E... finished Cgigas_v9_gene--ID
In [44]:
!python {sqls}fetchdata.py \
-s "SELECT * \
FROM [sr320@washington.edu].[Cgigas_v9_gene--ID]md \
left join \
[sr320@washington.edu].[qDOD_Cgigas_GOslim_DISTINCT]go on md.Column10=go.CGI_ID" \
-f tsv \
-o /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID--GOslim.tab
In [45]:
!head /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID--GOslim.tab
In [47]:
!tail -3 /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID--GOslim.tab
In [79]:
!grep 'DNA metabolism\|RNA metabolism\|protein metabolism' \
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID--GOslim.tab | head
In [80]:
%%bash
grep --color 'cell-cell signaling\|signal transduction\|cell adhesion' \
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID--GOslim.tab \
| grep -v "signal transduction activity F" \
| head
C17316 GLEAN mRNA 30 257 0.555898 + . ID=CGI_10000003; CGI_10000003 CGI_10000003 signal transduction P C20480 GLEAN mRNA 367 1037 0.999572 - . ID=CGI_10000032; CGI_10000032 CGI_10000032 signal transduction P C20578 GLEAN mRNA 699 950 0.555898 + . ID=CGI_10000034; CGI_10000034 CGI_10000034 signal transduction P C22046 GLEAN mRNA 98 1281 1 + . ID=CGI_10000069; CGI_10000069 CGI_10000069 cell adhesion P C22046 GLEAN mRNA 98 1281 1 + . ID=CGI_10000069; CGI_10000069 CGI_10000069 signal transduction P C22798 GLEAN mRNA 433 1785 1 + . ID=CGI_10000088; CGI_10000088 CGI_10000088 signal transduction P C23676 GLEAN mRNA 34 2210 1 + . ID=CGI_10000145; CGI_10000145 CGI_10000145 signal transduction P scaffold1370 GLEAN mRNA 642 1238 1 - . ID=CGI_10000165; CGI_10000165 CGI_10000165 signal transduction P scaffold1370 GLEAN mRNA 1243 2469 0.999414 - . ID=CGI_10000166; CGI_10000166 CGI_10000166 signal transduction P C24232 GLEAN mRNA 589 2415 1 - . ID=CGI_10000183; CGI_10000183 CGI_10000183 signal transduction P
In [87]:
#QC
!grep 'DNA metabolism\|RNA metabolism\|protein metabolism' \
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID--GOslim.tab \
| cut -f 12 | sort | uniq -c
666 DNA metabolism 2452 RNA metabolism 3737 protein metabolism
In [88]:
#QC
!grep 'cell-cell signaling\|signal transduction\|cell adhesion' \
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID--GOslim.tab \
| grep -v "signal transduction activity F" \
| cut -f 12 | sort | uniq -c
1069 cell adhesion 478 cell-cell signaling 3001 signal transduction
In [94]:
!grep 'DNA metabolism\|RNA metabolism\|protein metabolism' \
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID--GOslim.tab \
| cut -f 1,2,3,4,5,6,7,8,9 > /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff
In [95]:
!grep 'cell-cell signaling\|signal transduction\|cell adhesion' \
/Users/sr320/data-genomic/tentacle/Cgigas_v9_gene--ID--GOslim.tab \
| grep -v "signal transduction activity F" \
| cut -f 1,2,3,4,5,6,7,8,9 > /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff
In [ ]: