DNA methylation of Oyster Sperm based on Genomic Features¶
methratio file in SQLShare https://sqlshare.escience.washington.edu/sqlshare#s=query/sr320%2540washington.edu/clean_BiGo_methratio_v1
BiGO methratio GFF¶
SELECT
chr as seqname,
'methratio' as source,
'CpG' as feature,
pos as start,
pos + 1 as [end],
ratio as score,
strand,
'.' as frame,
'.' as attribute
FROM [sr320@washington.edu].[clean_BiGo_methratio_v1]
where
context like '__CG_'
and
CT_Count >= 5
In [13]:
# ie python fetchdata.py -d "[sr320@washington.edu].[BiGO_Methylation_oysterv9_GFF]" -f tsv -o /Volumes/web/cnidarian/BiGO_Methylation10x_oysterv9.gff
# running on commandline because cannot get to work in IPython
In [15]:
#fetchdata failed
In [16]:
#Dowloaded csv
# should be same as
!head /Volumes/web/cnidarian/BiGO_Methylation5x_oysterv9.gff
In [17]:
!wc /Volumes/web/cnidarian/BiGO_Methylation5x_oysterv9.gff
7648305 68834745 402812022 /Volumes/web/cnidarian/BiGO_Methylation5x_oysterv9.gff
In [1]:
!head /Volumes/web/cnidarian/BiGo_methratio_boop.gff
In [21]:
!wc /Volumes/web/cnidarian/BiGo_methratio_boop.gff
7642817 68785353 378876013 /Volumes/web/cnidarian/BiGo_methratio_boop.gff
In [125]:
from pandas import *
# read data from data file into a pandas DataFrame
BiGOboop = read_csv("http://eagle.fish.washington.edu/cnidarian/BiGo_methratio_GFF_boop.csv", # name of the data file
sep=",", # what character separates each column?
na_values=["", " "]) # what values should be considered "blank" values?
In [126]:
BiGOboop['score'].hist(bins=50);
#Axis limits are changed using the axis([xmin, xmax, ymin, ymax]) function.
plt.axis([0, 1, 0, 400000])
Out[126]:
[0, 1, 0, 400000]
CG motif¶
In [29]:
!head /Volumes/web/cnidarian/TJGR_oyster_v9_CGmotif.gff
##gff-version 2.0 ##date 2013-04-23 ##Type DNA scaffold360 scaffold360 fuzznuc misc_feature 60 61 2.000 + . Sequence "scaffold360.1" ; note "*pat pattern1" scaffold360 fuzznuc misc_feature 96 97 2.000 + . Sequence "scaffold360.2" ; note "*pat pattern1" scaffold360 fuzznuc misc_feature 120 121 2.000 + . Sequence "scaffold360.3" ; note "*pat pattern1" scaffold360 fuzznuc misc_feature 187 188 2.000 + . Sequence "scaffold360.4" ; note "*pat pattern1" ##gff-version 2.0 ##date 2013-04-23 ##Type DNA scaffold18356
In [30]:
!fgrep -c "fuzznuc" /Volumes/web/cnidarian/TJGR_oyster_v9_CGmotif.gff
5625744
In [31]:
!head /Volumes/web/cnidarian/TJGR_oyster_v9_CG.gff
##gff-version 3 ##sequence-region scaffold360 1 280 #!Date 2013-04-23 #!Type DNA #!Source-version EMBOSS 6.5.7.0 scaffold360 fuzznuc nucleotide_motif 60 61 2 + . ID=scaffold360.1;note=*pat pattern:CG scaffold360 fuzznuc nucleotide_motif 96 97 2 + . ID=scaffold360.2;note=*pat pattern:CG scaffold360 fuzznuc nucleotide_motif 120 121 2 + . ID=scaffold360.3;note=*pat pattern:CG scaffold360 fuzznuc nucleotide_motif 187 188 2 + . ID=scaffold360.4;note=*pat pattern:CG ##gff-version 3
In [32]:
!fgrep -c "fuzznuc" /Volumes/web/cnidarian/TJGR_oyster_v9_CG.gff
9978551
Confirming CG¶
In [7]:
#ran before
!head /Volumes/web/cnidarian/oyster_v9_CG_fuzznuc.output
######################################## # Program: fuzznuc # Rundate: Fri 19 Apr 2013 14:58:14 # Commandline: fuzznuc # -sequence oyster.v9.fa # -pattern CG # -outfile fuzznuc.output # Report_format: seqtable # Report_file: fuzznuc.output ########################################
In [9]:
!fgrep -c "+ pattern: " /Volumes/web/cnidarian/oyster_v9_CG_fuzznuc.output
9978551
In [10]:
!fgrep -c "nucleotide_motif" /Volumes/web/bivalvia/wholegenomefiles_MBDbsSeq_gill/gffs/TJGR_oyster_v9_CG.gff
9978551
CG Content in all Features¶
In [78]:
!intersectbed -a /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_exon.gff -b /Volumes/web/cnidarian/TJGR_oyster_v9_CG.gff -c > /Volumes/web/cnidarian/TGR_intersectbed_CDS_v9_CGmotif.txt
In [80]:
!intersectbed -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_exon.gff -a /Volumes/web/cnidarian/TJGR_oyster_v9_CG.gff > /Volumes/web/cnidarian/TGR_CGmotif_intersect_exon.txt
In [37]:
!head /Volumes/web/cnidarian/TGR_intersectbed_CDS_v9_CGmotif.txt
C16582 GLEAN CDS 35 385 . - 0 Parent=CGI_10000001; 14 C17212 GLEAN CDS 31 363 . + 0 Parent=CGI_10000002; 6 C17316 GLEAN CDS 30 257 . + 0 Parent=CGI_10000003; 10 C17476 GLEAN CDS 104 257 . - 0 Parent=CGI_10000004; 5 C17476 GLEAN CDS 34 74 . - 2 Parent=CGI_10000004; 2 C17998 GLEAN CDS 196 387 . - 0 Parent=CGI_10000005; 11 C18346 GLEAN CDS 174 551 . + 0 Parent=CGI_10000009; 29 C18428 GLEAN CDS 286 546 . - 0 Parent=CGI_10000010; 24 C18964 GLEAN CDS 203 658 . - 0 Parent=CGI_10000011; 9 C18980 GLEAN CDS 30 674 . + 0 Parent=CGI_10000012; 56
In [76]:
!wc /Volumes/web/cnidarian/TGR_intersectbed_CDS_v9_CGmotif.txt
196691 1966910 12782384 /Volumes/web/cnidarian/TGR_intersectbed_CDS_v9_CGmotif.txt
In [43]:
cat /Volumes/web/cnidarian/TGR_intersectbed_CDS_v9_CGmotif.txt | awk -F"\t" '{ sum+=$10} END {print sum}'
1134622
In [81]:
!head /Volumes/web/cnidarian/TGR_CGmotif_intersect_exon.txt
scaffold350 fuzznuc nucleotide_motif 1161 1162 2 + . ID=scaffold350.25;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 1183 1184 2 + . ID=scaffold350.26;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 1200 1201 2 + . ID=scaffold350.27;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 1252 1253 2 + . ID=scaffold350.28;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 1267 1268 2 + . ID=scaffold350.29;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 1287 1288 2 + . ID=scaffold350.30;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 1331 1332 2 + . ID=scaffold350.31;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 1416 1417 2 + . ID=scaffold350.32;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 1439 1440 2 + . ID=scaffold350.33;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 1480 1481 2 + . ID=scaffold350.34;note=*pat pattern:CG
In [82]:
!wc /Volumes/web/cnidarian/TGR_CGmotif_intersect_exon.txt
1134622 11346220 111446503 /Volumes/web/cnidarian/TGR_CGmotif_intersect_exon.txt
In [86]:
## CG intersect intron
!intersectbed -a /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_CG.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_intron.gff > /Volumes/web/cnidarian/TGR_intersectbed_intron_v9_CG.gff
In [87]:
!wc /Volumes/web/cnidarian/TGR_intersectbed_intron_v9_CG.gff
2886432 28864320 283836671 /Volumes/web/cnidarian/TGR_intersectbed_intron_v9_CG.gff
In [88]:
## CG intersect TE
!intersectbed -a /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_CG.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE.gff > /Volumes/web/cnidarian/TGR_intersectbed_CG_TE.gff
In [89]:
!wc /Volumes/web/cnidarian/TGR_intersectbed_CG_TE.gff
832569 8325690 81351343 /Volumes/web/cnidarian/TGR_intersectbed_CG_TE.gff
In [90]:
## CG intersect Promoter
!intersectbed -a /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_CG.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff > /Volumes/web/cnidarian/TGR_intersectbed_CG_prom.gff
In [91]:
!wc /Volumes/web/cnidarian/TGR_intersectbed_CG_prom.gff
616223 6162230 60375672 /Volumes/web/cnidarian/TGR_intersectbed_CG_prom.gff
In [1]:
!head /Volumes/web/cnidarian/TGR_intersectbed_CG_prom.gff
scaffold350 fuzznuc nucleotide_motif 3250 3251 2 + . ID=scaffold350.82;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 3266 3267 2 + . ID=scaffold350.83;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 3299 3300 2 + . ID=scaffold350.84;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 3316 3317 2 + . ID=scaffold350.85;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 3384 3385 2 + . ID=scaffold350.86;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 3474 3475 2 + . ID=scaffold350.87;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 3497 3498 2 + . ID=scaffold350.88;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 3505 3506 2 + . ID=scaffold350.89;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 3608 3609 2 + . ID=scaffold350.90;note=*pat pattern:CG scaffold350 fuzznuc nucleotide_motif 3622 3623 2 + . ID=scaffold350.91;note=*pat pattern:CG
In [92]:
## CG intersect Promoter
!intersectbed -a /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_CG.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_COMP_gene_prom_TE.bed > /Volumes/web/cnidarian/TGR_intersectbed_CG_other.gff
In [93]:
!wc /Volumes/web/cnidarian/TGR_intersectbed_CG_other.gff
5121236 51212360 499097421 /Volumes/web/cnidarian/TGR_intersectbed_CG_other.gff
mCG per feature¶
SELECT
chr as seqname,
'methratio' as source,
'CpG' as feature,
pos as start,
pos + 1 as [end],
ratio as score,
strand,
'.' as frame,
'.' as attribute
FROM [sr320@washington.edu].[clean_BiGo_methratio_v1]
where
context like '__CG_'
and
CT_Count >= 5
and
ratio >= 0.5
In [103]:
!head /Volumes/web/cnidarian/BiGo_methratio_mCG.csv
In [104]:
!wc /Volumes/web/cnidarian/BiGo_methratio_mCG.csv
1159721 1159721 59922755 /Volumes/web/cnidarian/BiGo_methratio_mCG.csv
In [105]:
!tr ',' "\t" </Volumes/web/cnidarian/BiGo_methratio_mCG.csv> /Volumes/web/cnidarian/BiGo_methratio_mCG.gff
In [106]:
!head /Volumes/web/cnidarian/BiGo_methratio_mCG.gff
In [107]:
!tail -n +2 /Volumes/web/cnidarian/BiGo_methratio_mCG.gff > /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff
In [109]:
!head /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff
In [110]:
###mCG intersects with genome features
In [111]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_exon.gff > /Volumes/web/cnidarian/TGR_mCG_intersect_exon.gff
In [121]:
!wc /Volumes/web/cnidarian/TGR_mCG_intersect_exon.gff
343032 3087288 17692941 /Volumes/web/cnidarian/TGR_mCG_intersect_exon.gff
In [113]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_intron.gff > /Volumes/web/cnidarian/TGR_mCG_intersect_intron.gff
In [114]:
!wc /Volumes/web/cnidarian/TGR_mCG_intersect_intron.gff
518734 4668606 26896216 /Volumes/web/cnidarian/TGR_mCG_intersect_intron.gff
In [115]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE.gff > /Volumes/web/cnidarian/TGR_mCG_intersect_TE.gff
In [116]:
!wc /Volumes/web/cnidarian/TGR_mCG_intersect_TE.gff
38532 346788 1994156 /Volumes/web/cnidarian/TGR_mCG_intersect_TE.gff
In [117]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff > /Volumes/web/cnidarian/TGR_mCG_intersect_prom.gff
In [118]:
!wc /Volumes/web/cnidarian/TGR_mCG_intersect_prom.gff
45241 407169 2336384 /Volumes/web/cnidarian/TGR_mCG_intersect_prom.gff
In [119]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_COMP_gene_prom_TE.bed > /Volumes/web/cnidarian/TGR_mCG_intersect_other.gff
In [120]:
!wc /Volumes/web/cnidarian/TGR_mCG_intersect_other.gff
277587 2498283 14287492 /Volumes/web/cnidarian/TGR_mCG_intersect_other.gff
| Feature | No. Features | CG | mCG | NmCG |
|---|---|---|---|---|
| Exons | 196691 | 1134622 | 343032 | NmCG |
| Intron | 176049 | 2886432 | 518734 | NmCG |
| TE | 119786 | 832569 | 38532 | NmCG |
| Promoter | 28023 | 616223 | 45241 | NmCG |
| Other | 81736 | 5121236 | 277587 | NmCG |
In [99]:
sum(1134622+2886432+616223+5121236)
Out[99]:
9758513
Histogram for all features¶
In [5]:
!tail -n +2 /Volumes/web/cnidarian/BiGo_methratio_boop.gff > /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff
In [6]:
!head /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff
In [7]:
!wc /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff
7642816 68785344 378875950 /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff
In [8]:
!wc /Volumes/web/cnidarian/BiGo_methratio_boop.gff
7642817 68785353 378876013 /Volumes/web/cnidarian/BiGo_methratio_boop.gff
In [11]:
!intersectbed -a /Volumes/web/cnidarian/oyster.v9.glean.final.rename.CDS.gff -b /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff -c > /Volumes/web/cnidarian/BiGo_CDS_v9_intersect_methratio_boop.txt
In [10]:
!head /Volumes/web/cnidarian/BiGo_CDS_v9_intersect_methratio_boop.txt
C16582 GLEAN CDS 35 385 . - 0 Parent=CGI_10000001; 0 C17212 GLEAN CDS 31 363 . + 0 Parent=CGI_10000002; 4 C17316 GLEAN CDS 30 257 . + 0 Parent=CGI_10000003; 9 C17476 GLEAN CDS 104 257 . - 0 Parent=CGI_10000004; 5 C17476 GLEAN CDS 34 74 . - 2 Parent=CGI_10000004; 1 C17998 GLEAN CDS 196 387 . - 0 Parent=CGI_10000005; 11 C18346 GLEAN CDS 174 551 . + 0 Parent=CGI_10000009; 0 C18428 GLEAN CDS 286 546 . - 0 Parent=CGI_10000010; 0 C18964 GLEAN CDS 203 658 . - 0 Parent=CGI_10000011; 9 C18980 GLEAN CDS 30 674 . + 0 Parent=CGI_10000012; 0
In [12]:
#wrong order
BiGo methratio intersect Exon¶
In [35]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_exon.gff > /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_CDS_b.gff
In [36]:
!wc /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_CDS_b.gff
1028202 9253818 51391228 /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_CDS_b.gff
In [37]:
!head /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_CDS_b.gff
In [38]:
from pandas import *
# read data from data file into a pandas DataFrame
CDSmr = read_table("/Volumes/web/cnidarian/BiGo_methratio_boop_intersect_CDS_b.gff", # name of the data file
#sep="\t", # what character separates each column?
#na_values=["", " "], # what values should be considered "blank" values?
header=None)
In [129]:
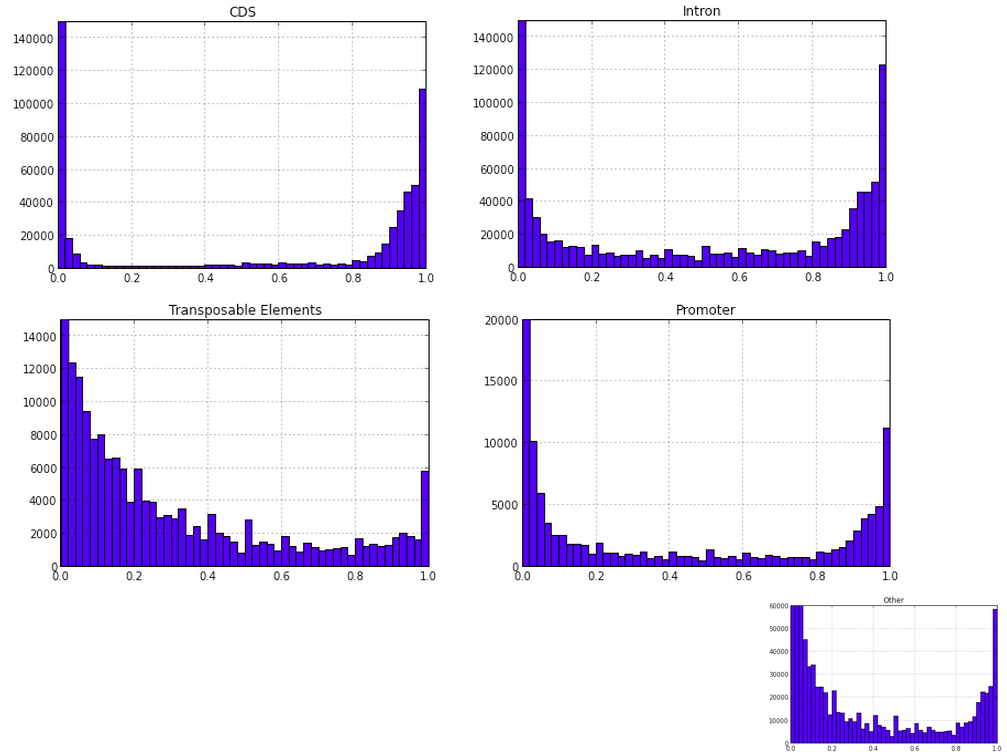
CDSmr[5].hist(bins=50);
#Axis limits are changed using the axis([xmin, xmax, ymin, ymax]) function.
plt.axis([0, 1, 0, 150000]);
plt.title('CDS');
In [70]:
# pandas density plot
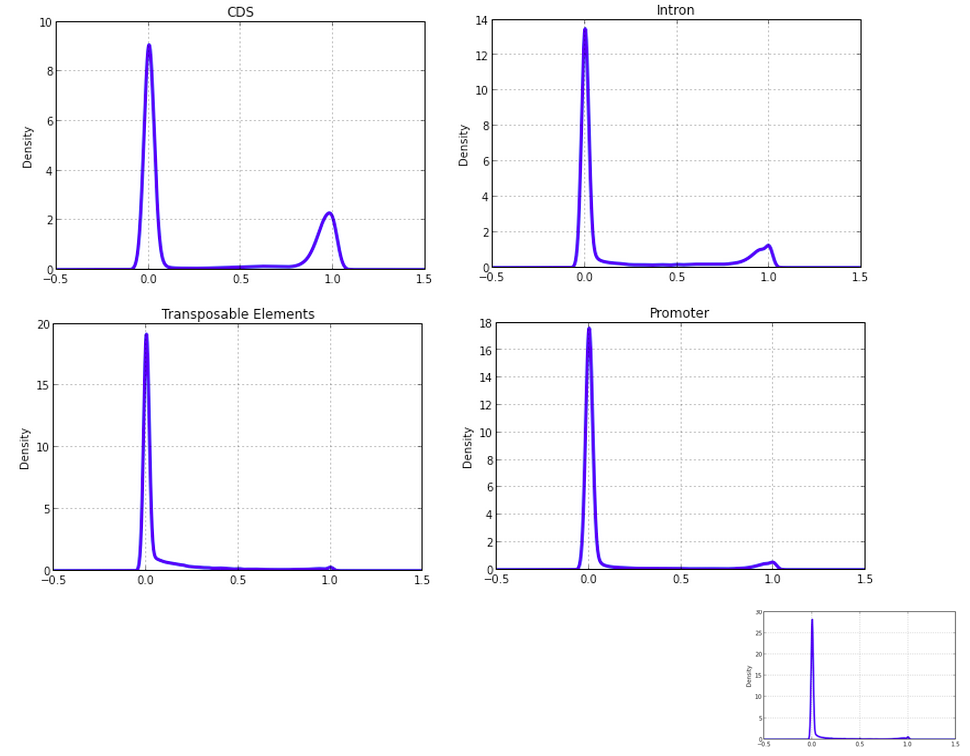
CDSmr[5].plot(kind='kde', linewidth=3);
plt.title('CDS')
Out[70]:
<matplotlib.text.Text at 0x1069706d0>
BiGo methratio intersect Promoter¶
In [52]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff > /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_promoter.gff
In [53]:
!head /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_promoter.gff
In [54]:
from pandas import *
# read data from data file into a pandas DataFrame
Promr = read_table("/Volumes/web/cnidarian/BiGo_methratio_boop_intersect_promoter.gff", # name of the data file
#sep="\t", # what character separates each column?
#na_values=["", " "], # what values should be considered "blank" values?
header=None)
In [132]:
Promr[5].hist(bins=50);
#Axis limits are changed using the axis([xmin, xmax, ymin, ymax]) function.
plt.axis([0, 1, 0, 20000]);
plt.title('Promoter');
In [139]:
# pandas density plot
Promr[5].plot(kind='kde', linewidth=3);
plt.title('Promoter');
BiGo methratio intersect TE¶
In [49]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE.gff > /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_TE.gff
In [50]:
!head /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_TE.gff
In [56]:
from pandas import *
# read data from data file into a pandas DataFrame
TEmr = read_table("/Volumes/web/cnidarian/BiGo_methratio_boop_intersect_TE.gff", # name of the data file
#sep="\t", # what character separates each column?
#na_values=["", " "], # what values should be considered "blank" values?
header=None)
In [63]:
TEmr[5].hist(bins=50);
#Axis limits are changed using the axis([xmin, xmax, ymin, ymax]) function.
plt.axis([0, 1, 0, 15000]);
plt.title('Transposable Elements');
In [140]:
# pandas density plot
TEmr[5].plot(kind='kde', linewidth=3);
plt.title('Transposable Elements');
BiGO methration intersect intron¶
In [60]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_intron.gff > /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_intron.gff
In [61]:
from pandas import *
# read data from data file into a pandas DataFrame
Intronmr = read_table("/Volumes/web/cnidarian/BiGo_methratio_boop_intersect_intron.gff", # name of the data file
#sep="\t", # what character separates each column?
#na_values=["", " "], # what values should be considered "blank" values?
header=None)
In [136]:
Intronmr[5].hist(bins=50);
#Axis limits are changed using the axis([xmin, xmax, ymin, ymax]) function.
plt.axis([0, 1, 0, 150000]);
plt.title('Intron');
In [141]:
# pandas density plot
Intronmr[5].plot(kind='kde', linewidth=3);
plt.title('Intron');
BiGO methratio intersect 'other'¶
In [66]:
!intersectbed -a /Volumes/web/cnidarian/BiGo_methratio_boop_c.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_COMP_gene_prom_TE.bed > /Volumes/web/cnidarian/BiGo_methratio_boop_intersect_other.gff
In [67]:
from pandas import *
# read data from data file into a pandas DataFrame
othermr = read_table("/Volumes/web/cnidarian/BiGo_methratio_boop_intersect_other.gff", # name of the data file
#sep="\t", # what character separates each column?
#na_values=["", " "], # what values should be considered "blank" values?
header=None)
In [138]:
othermr[5].hist(bins=50);
#Axis limits are changed using the axis([xmin, xmax, ymin, ymax]) function.
plt.axis([0, 1, 0, 60000]);
plt.title('Other');
In [101]:
# pandas density plot
othermr[5].plot(kind='kde', linewidth=3);
plt.title('Other');
Gene Level Methylation¶
In [1]:
#For each gene, need to get number of CG and Number of methylated CG
#methylated CG defined as 5x 50%
In [2]:
#number of CGs per gene
!intersectbed -c -a /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_gene.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_CG.gff > /Volumes/web/cnidarian/TJGR_gene_CGcount.txt
In [3]:
!head /Volumes/web/cnidarian/TJGR_gene_CGcount.txt
C16582 GLEAN mRNA 35 385 0.555898 - . ID=CGI_10000001; 14 C17212 GLEAN mRNA 31 363 0.999572 + . ID=CGI_10000002; 6 C17316 GLEAN mRNA 30 257 0.555898 + . ID=CGI_10000003; 10 C17476 GLEAN mRNA 34 257 0.998947 - . ID=CGI_10000004; 7 C17998 GLEAN mRNA 196 387 1 - . ID=CGI_10000005; 11 C18346 GLEAN mRNA 174 551 1 + . ID=CGI_10000009; 29 C18428 GLEAN mRNA 286 546 0.555898 - . ID=CGI_10000010; 24 C18964 GLEAN mRNA 203 658 0.999572 - . ID=CGI_10000011; 9 C18980 GLEAN mRNA 30 674 0.555898 + . ID=CGI_10000012; 56 C19100 GLEAN mRNA 160 681 0.999955 - . ID=CGI_10000013; 12
In [6]:
!wc /Volumes/web/cnidarian/TJGR_gene_CGcount.txt
28027 280270 1915935 /Volumes/web/cnidarian/TJGR_gene_CGcount.txt
In [41]:
#BiGO methylaiton file
!head /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff
In [42]:
!tail /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff
In [7]:
#number of mCGs per gene
!intersectbed -c -a /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_gene.gff -b /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff > /Volumes/web/cnidarian/BiGO_gene_mCGcount.txt
In [8]:
!head /Volumes/web/cnidarian/BiGO_gene_mCGcount.txt
C16582 GLEAN mRNA 35 385 0.555898 - . ID=CGI_10000001; 0 C17212 GLEAN mRNA 31 363 0.999572 + . ID=CGI_10000002; 0 C17316 GLEAN mRNA 30 257 0.555898 + . ID=CGI_10000003; 0 C17476 GLEAN mRNA 34 257 0.998947 - . ID=CGI_10000004; 0 C17998 GLEAN mRNA 196 387 1 - . ID=CGI_10000005; 0 C18346 GLEAN mRNA 174 551 1 + . ID=CGI_10000009; 0 C18428 GLEAN mRNA 286 546 0.555898 - . ID=CGI_10000010; 0 C18964 GLEAN mRNA 203 658 0.999572 - . ID=CGI_10000011; 0 C18980 GLEAN mRNA 30 674 0.555898 + . ID=CGI_10000012; 0 C19100 GLEAN mRNA 160 681 0.999955 - . ID=CGI_10000013; 8
In [39]:
!tail /Volumes/web/cnidarian/BiGO_gene_mCGcount.txt
scaffold22 GLEAN mRNA 1710615 1726578 0.998897 + . ID=CGI_10028929; 59 scaffold22 GLEAN mRNA 1728744 1730768 0.999999 - . ID=CGI_10028930; 0 scaffold22 GLEAN mRNA 1740746 1762025 0.648316 + . ID=CGI_10028931; 175 scaffold22 GLEAN mRNA 1748291 1749451 1 - . ID=CGI_10028932; 11 scaffold22 GLEAN mRNA 1763621 1771882 1 + . ID=CGI_10028933; 46 scaffold22 GLEAN mRNA 1784983 1800181 0.865666 - . ID=CGI_10028934; 0 scaffold22 GLEAN mRNA 1819528 1821088 0.988961 + . ID=CGI_10028935; 0 scaffold22 GLEAN mRNA 1863347 1863687 0.647996 + . ID=CGI_10028937; 0 scaffold22 GLEAN mRNA 1863760 1864161 0.544455 + . ID=CGI_10028938; 0 scaffold22 GLEAN mRNA 1869336 1885890 0.999933 - . ID=CGI_10028939; 0
In [40]:
!wc /Volumes/web/cnidarian/BiGO_gene_mCGcount.txt
28027 280270 1888975 /Volumes/web/cnidarian/BiGO_gene_mCGcount.txt
In [43]:
#join
#going for ugly excel cut and paste
In [ ]:
#sneak peek at percent methylation per gene (when CG count >/= 10)
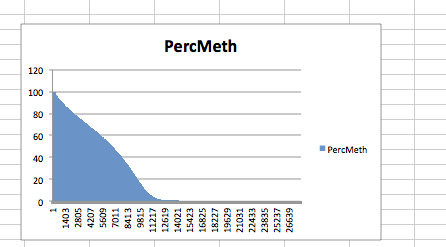
In [45]:
#only genes wiht >/= 10 cgs
!head /Volumes/web/cnidarian/BiGo_gene_PerMeth.txt
GeneID CG mCG PercMeth CGI_10000309 10 10 100 CGI_10002318 10 10 100 CGI_10003667 10 10 100 CGI_10003855 10 10 100 CGI_10004087 10 10 100 CGI_10005035 10 10 100 CGI_10007691 10 10 100 CGI_10009829 10 10 100 CGI_10014371 10 10 100
In [44]:
#now need to examine percent methylation versus gene expression
sqlshare code to get expression
SELECT *
FROM [sr320@washington.edu].[table_BiGo_gene_PerMeth.txt]gene
left join
[sr320@washington.edu].[BiGo_RNAseq_genes]exp
on
gene.GeneID = exp.["Feature ID"]
In [47]:
!head /Volumes/web/cnidarian/BiGo_gene_pmeth_expression.csv
GeneID,CG,mCG,PercMeth,expression,genelength,uniquecount,totalcounts,rpkm CGI_10000001,14,0,0,15.319,351,0,14,15.319 CGI_10000003,10,0,0,0,228,0,0,0 CGI_10000005,11,0,0,0,192,0,0,0 CGI_10000009,29,0,0,8.129,378,8,8,8.129 CGI_10000010,24,0,0,103.009,261,0,70,103.009 CGI_10000012,56,0,0,0.595,645,1,1,0.595 CGI_10000014,21,0,0,338.241,243,0,214,338.241 CGI_10000016,12,0,0,0,252,0,0,0 CGI_10000017,14,0,0,0,447,0,0,0
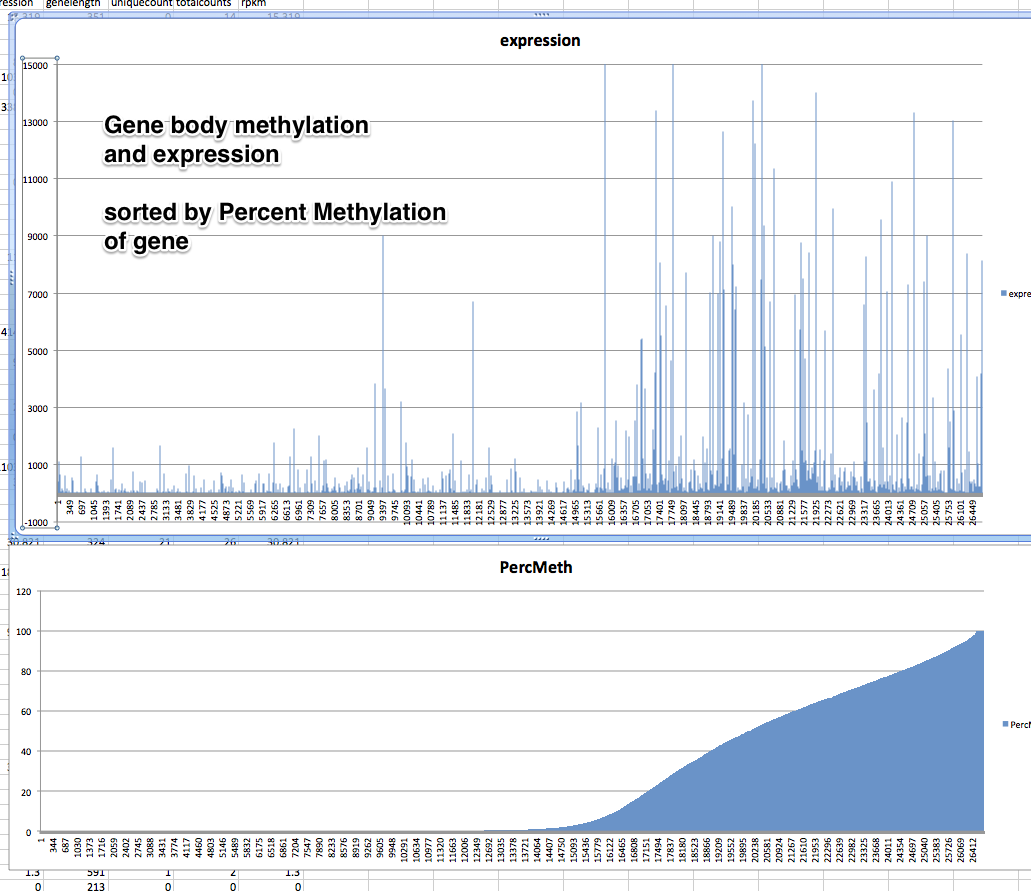
Promoter Methylation per gene¶
In [12]:
#will use promoter track that does not overlap genebodies
!wc /Volumes/web/cnidarian/TJGR_prom_subtract_gene1.gff
26706 240354 1803061 /Volumes/web/cnidarian/TJGR_prom_subtract_gene1.gff
In [17]:
!wc /Volumes/web/cnidarian/TJGR_prom_subgene_CGcount.txt
26706 267060 1877877 /Volumes/web/cnidarian/TJGR_prom_subgene_CGcount.txt
In [34]:
#post tw/excel modding
!head /Volumes/web/cnidarian/TJGR_prom_subgene_CGcount.txt
C16582 flankbed promoter 386 395 . - . CGI_10000001 0 C17212 flankbed promoter 1 30 . + . CGI_10000002 0 C17316 flankbed promoter 1 29 . + . CGI_10000003 0 C17476 flankbed promoter 258 491 . - . CGI_10000004 10 C17998 flankbed promoter 388 559 . - . CGI_10000005 8 C18346 flankbed promoter 1 173 . + . CGI_10000009 21 C18428 flankbed promoter 547 611 . - . CGI_10000010 2 C18964 flankbed promoter 659 714 . - . CGI_10000011 2 C18980 flankbed promoter 1 29 . + . CGI_10000012 5 C19100 flankbed promoter 682 743 . - . CGI_10000013 2
In [16]:
!head /Volumes/web/cnidarian/BiGo_prom_subgene_CGcount.txt
0 0 0 0 0 0 0 0 0 0
crazy line skip issue
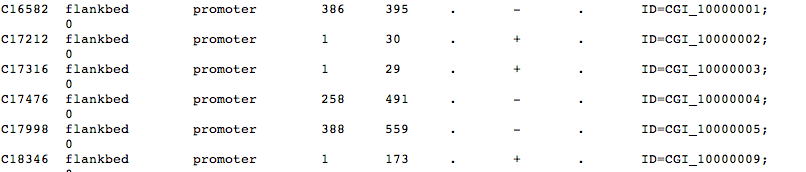
In [35]:
#post tw/excel modding
!head /Volumes/web/cnidarian/BiGo_prom_subgene_mCGcount.txt
C16582 flankbed promoter 386 395 . - . CGI_10000001 0 C17212 flankbed promoter 1 30 . + . CGI_10000002 0 C17316 flankbed promoter 1 29 . + . CGI_10000003 0 C17476 flankbed promoter 258 491 . - . CGI_10000004 0 C17998 flankbed promoter 388 559 . - . CGI_10000005 0 C18346 flankbed promoter 1 173 . + . CGI_10000009 0 C18428 flankbed promoter 547 611 . - . CGI_10000010 0 C18964 flankbed promoter 659 714 . - . CGI_10000011 0 C18980 flankbed promoter 1 29 . + . CGI_10000012 0 C19100 flankbed promoter 682 743 . - . CGI_10000013 0
In SQLSHare
Very close
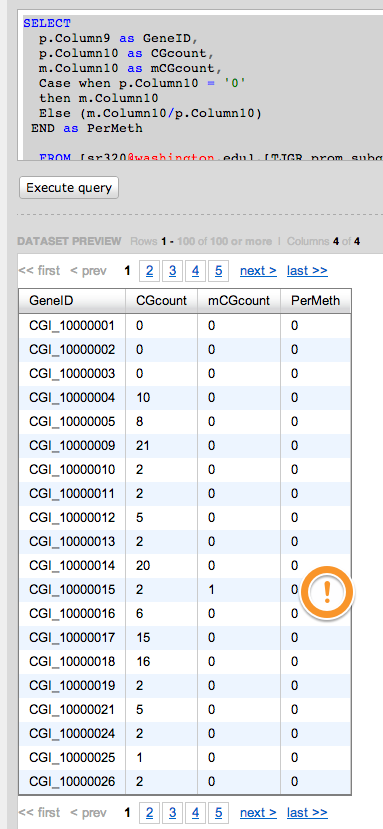
SELECT
p.Column9 as GeneID,
p.Column10 as CGcount,
m.Column10 as mCGcount,
Case when p.Column10 = '0'
then m.Column10
Else (m.Column10/p.Column10)
END as PerMeth
FROM [sr320@washington.edu].[TJGR_prom_subgene_CGcount.txt]p
LEFT Join
[sr320@washington.edu].[BiGo_prom_subgene_mCGcount.txt]m
on
p.Column9 = m.Column9
In [37]:
#give up into EXcel
!head /Volumes/web/cnidarian/BiGo_prom_subgene_PerMeth.txt
GeneID CGcount mCGcount Pmeth CGI_10001604 10 10 100 CGI_10020784 10 10 100 CGI_10021695 10 10 100 CGI_10021827 10 10 100 CGI_10021844 10 10 100 CGI_10021960 10 10 100 CGI_10011876 10 10 100 CGI_10009511 10 10 100 CGI_10006768 10 10 100
code
SELECT *
FROM [sr320@washington.edu].[BiGo_prom_subgene_PerMeth.txt]pr
left join
[sr320@washington.edu].[BiGo_RNAseq_genes]exp
on
pr.GeneID = exp.["Feature ID"]
In [1]:
#file manipulated in excel
!head /Volumes/web/cnidarian/BiGo_prom_subgene_pmeth_expression.txt
GeneID CGcount mCGcount Pmeth Exp CGI_10012330 27 0 0 35637.269 CGI_10001766 21 0 0 19885.868 CGI_10021069 19 0 0 17576.275 CGI_10012474 15 1 6.666666667 13030.698 CGI_10014767 42 0 0 12227.203 CGI_10026412 30 0 0 10903.164 CGI_10008493 18 1 5.555555556 10023.721 CGI_10024065 26 0 0 9939.849 CGI_10012002 13 9 69.23076923 9554.514
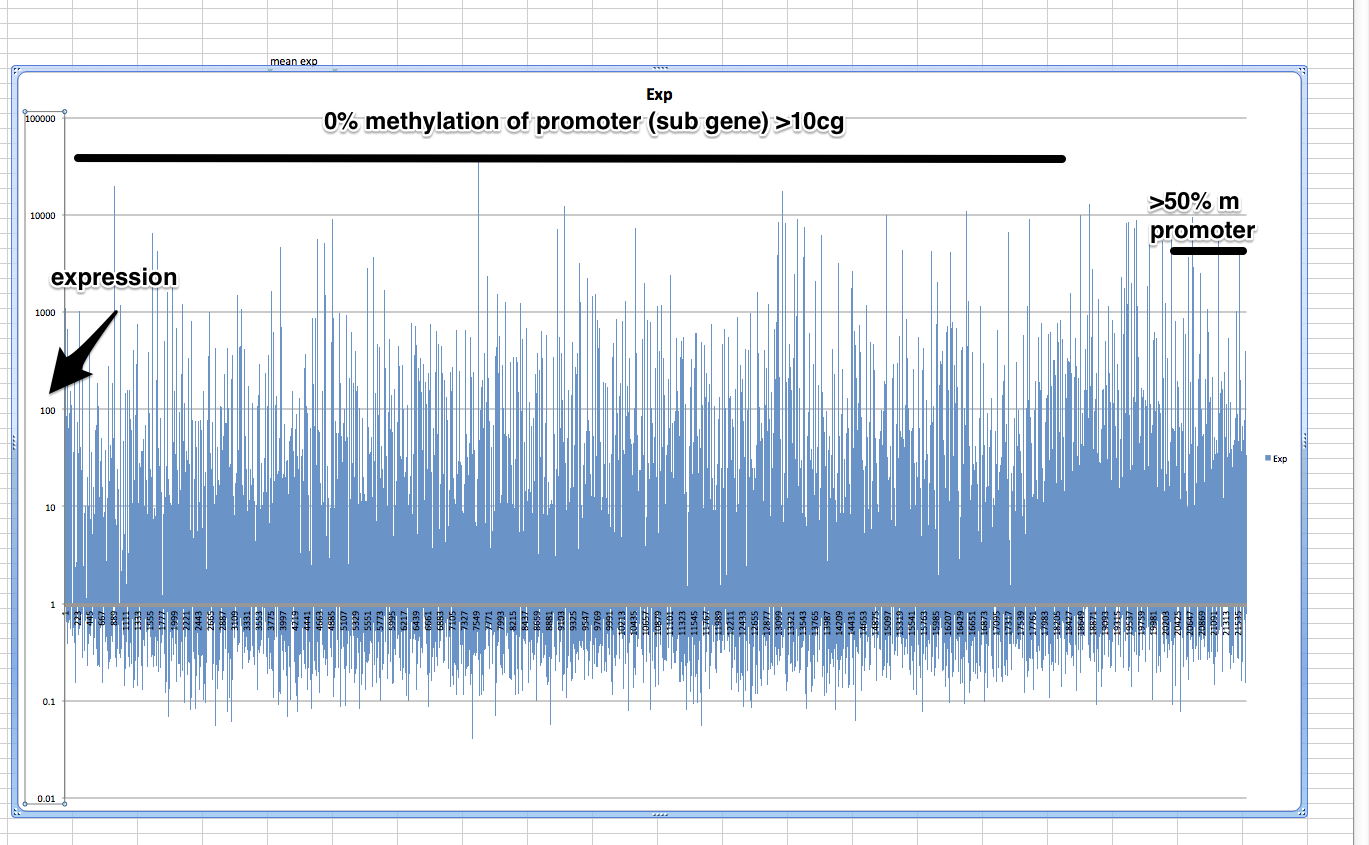
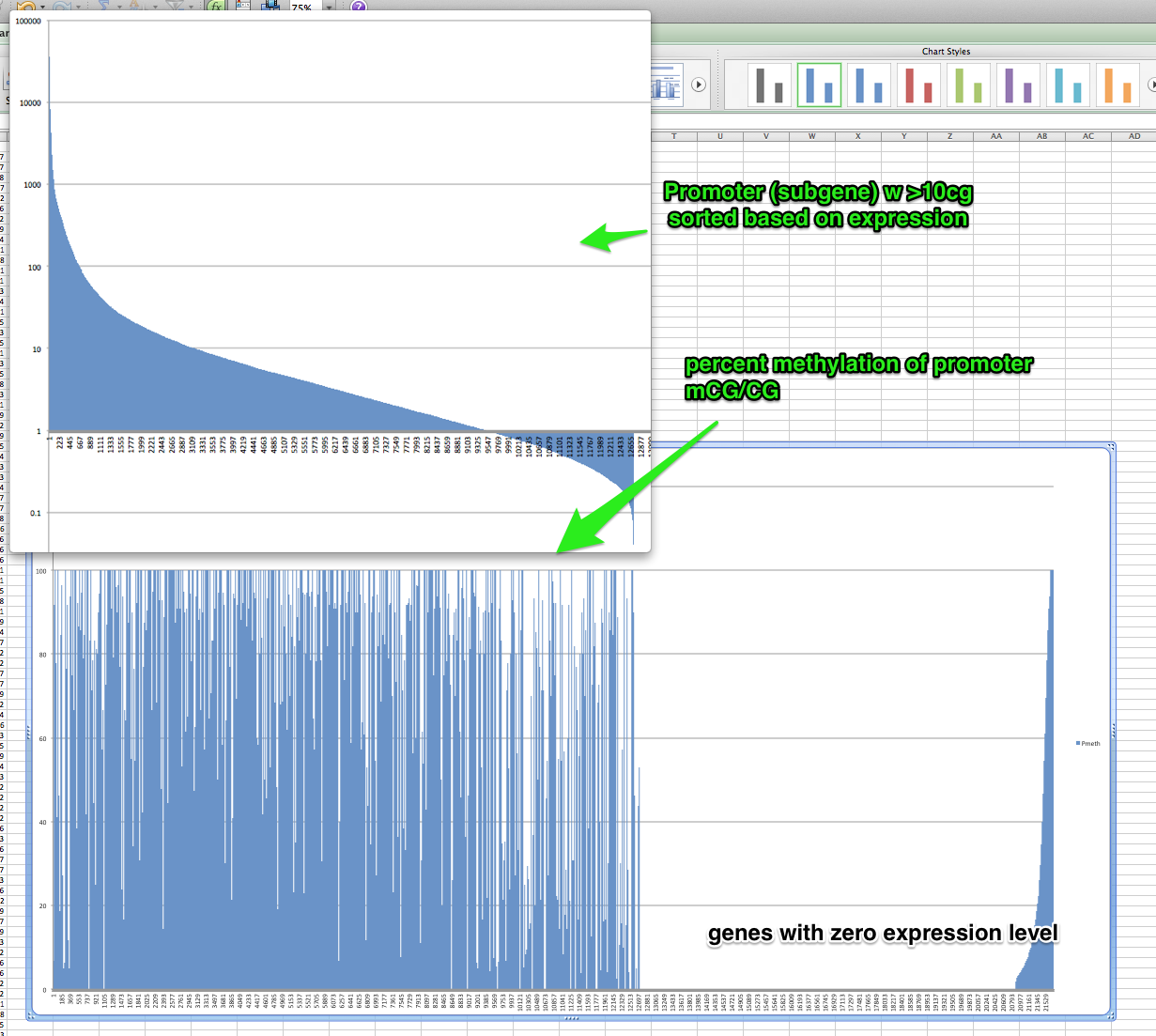
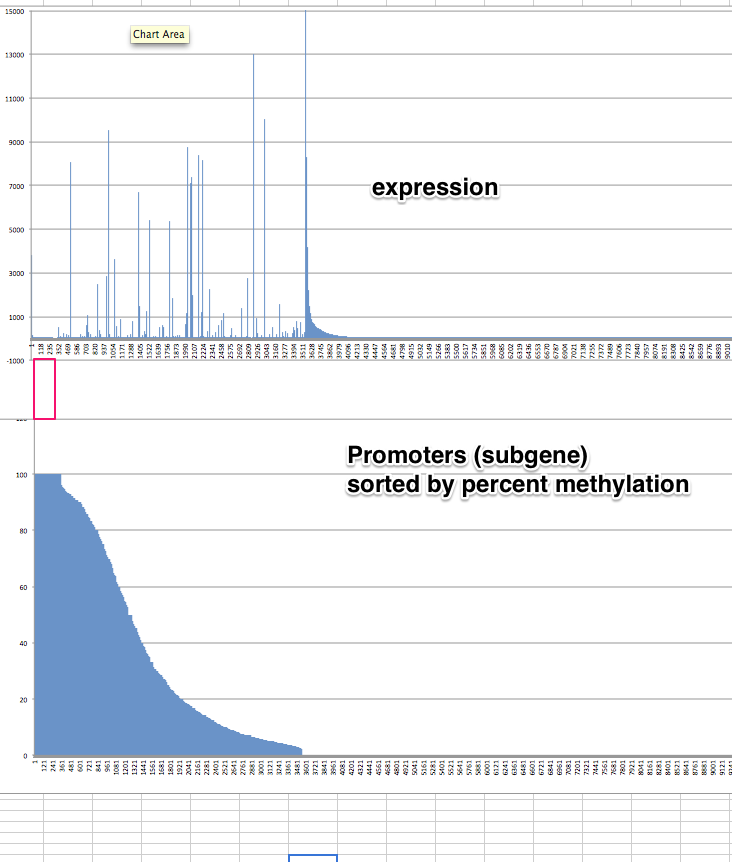
Closeup with random
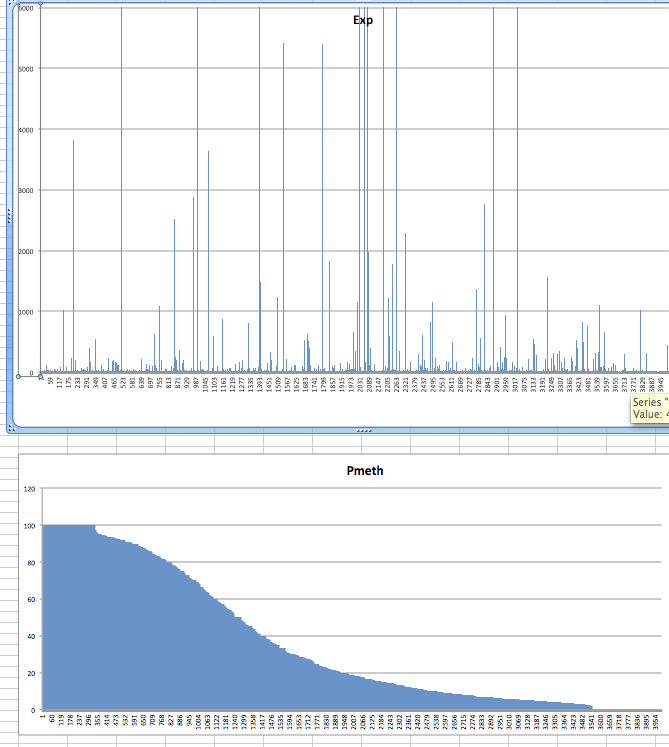 mm
mm
Promoter with CpG Islands methylation¶
In [ ]:
In [19]:
#absolute methylation
#using the conserved model
#Window size [100]:
#Minimum length of an island [200]:
#Minimum observed/expected [0.6]:
#Minimum percentage [50.]: 45
#promoter does not overlap gene
!intersectbed -c -a /Volumes/web/cnidarian/TJGR_prom_notgene_cpgIsland1u.gff -b /Volumes/web/cnidarian/BiGo_methratio_mCG_tail.gff > /Volumes/web/cnidarian/BiGo_pro_island1u_intersect_mCpG.txt
In [49]:
!head /Volumes/web/cnidarian/BiGo_pro_island1u_intersect_mCpG.txt
C18346 flankbed promoter 1 173 . + . CGI_10000009 0 C18428 flankbed promoter 547 611 . - . CGI_10000010 0 C19356 flankbed promoter 1 354 . + . CGI_10000014 0 C19532 flankbed promoter 602 843 . - . CGI_10000017 0 C20578 flankbed promoter 1 698 . + . CGI_10000034 0 C21046 flankbed promoter 813 1275 . - . CGI_10000042 0 C21254 flankbed promoter 1 669 . + . CGI_10000047 0 C21260 flankbed promoter 1176 1343 . - . CGI_10000048 0 C22036 flankbed promoter 1 947 . + . CGI_10000068 0 C22346 flankbed promoter 756 1755 . - . CGI_10000079 0
In [21]:
!wc /Volumes/web/cnidarian/BiGo_pro_island1u_intersect_mCpG.txt
2464 24640 170901 /Volumes/web/cnidarian/BiGo_pro_island1u_intersect_mCpG.txt
In [29]:
!intersectbed -c -a /Volumes/web/cnidarian/TJGR_prom_notgene_cpgIsland1u.gff -b /Volumes/web/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_CG.gff > /Volumes/web/cnidarian/TJGR_pro_island1u_intersect_CpG.txt
In [30]:
!wc /Volumes/web/cnidarian/TJGR_pro_island1u_intersect_CpG.txt
2464 24640 173098 /Volumes/web/cnidarian/TJGR_pro_island1u_intersect_CpG.txt
In [31]:
#geneexprssion level
!head /Volumes/web/cnidarian/BiGo_RNAseq_genes
"Feature ID" "Expression values" "Gene length" "Unique gene reads" "Total gene reads" "RPKM" CGI_10000780 0 1350 0 0 0 CGI_10000456 7.892 438 8 9 7.892 CGI_10000457 7.643 603 6 12 7.643 CGI_10000774 0 375 0 0 0 CGI_10000917 0 426 0 0 0 CGI_10000861 0 2004 0 0 0 CGI_10000994 16.913 1635 64 72 16.913 CGI_10000643 0.696 552 1 1 0.696 CGI_10000763 0 249 0 0 0
In [32]:
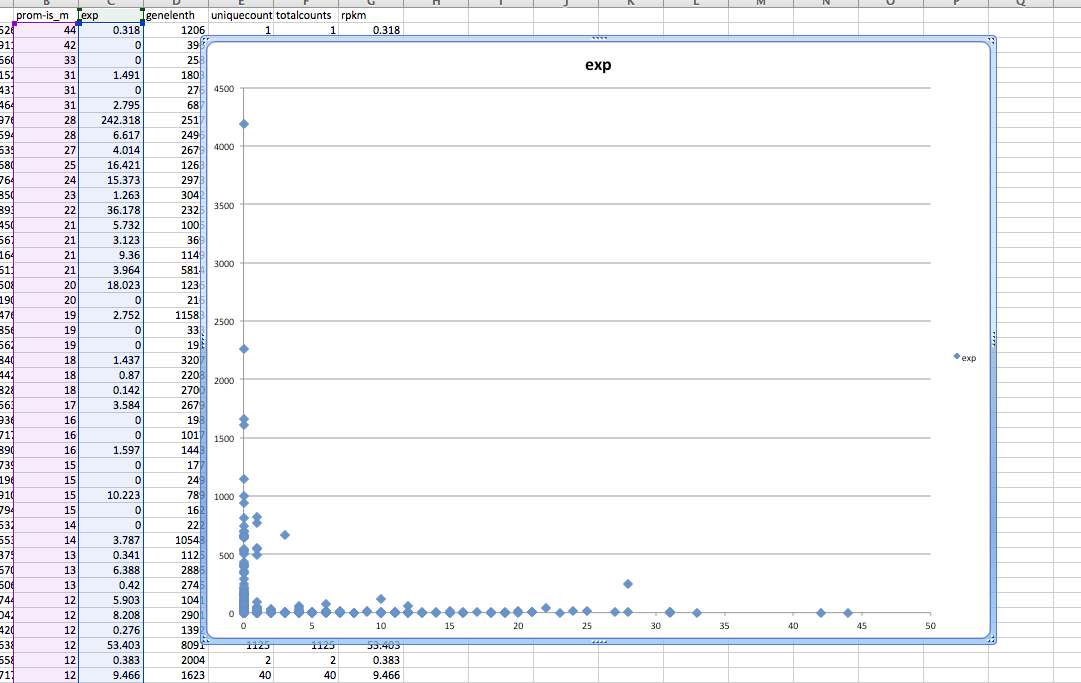
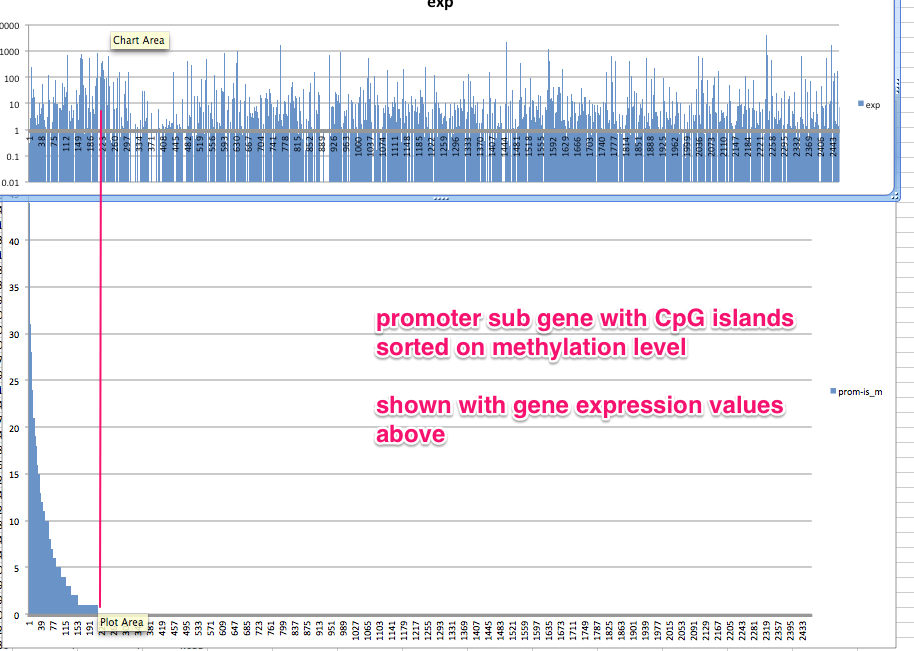
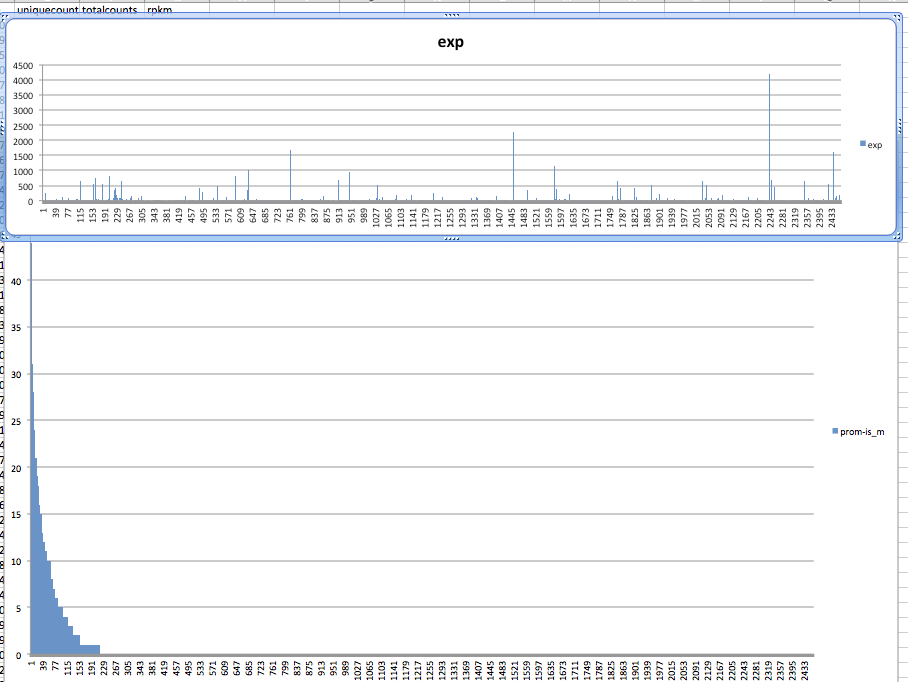
#lets plot absolute methylation versus expression
In [54]:
#simpled up methylation file
!wc /Volumes/web/cnidarian/BiGo_pro_island1u_intersect_mCpG_slm.csv
0 2464 37020 /Volumes/web/cnidarian/BiGo_pro_island1u_intersect_mCpG_slm.csv
code
SELECT *
FROM [sr320@washington.edu].[table_BiGo_pro_island1u_intersect_mCpG_slm.csv]pr_is
left join
[sr320@washington.edu].[BiGo_RNAseq_genes]exp
on
pr_is.Column1 = exp.["Feature ID"]
In [ ]: