In [1]:
from preamble import *
%matplotlib inline
Algorithm Chains and Pipelines¶
- train_test_split에 의하여 분할된 훈련 데이터 전체를 MinMaxScaler에 Fit을 함
In [2]:
from sklearn.svm import SVC
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import MinMaxScaler
# load and split the data
cancer = load_breast_cancer()
X_train, X_test, y_train, y_test = train_test_split(cancer.data, cancer.target, random_state=0)
# compute minimum and maximum on the training data
scaler = MinMaxScaler().fit(X_train)
In [3]:
# rescale the training data
X_train_scaled = scaler.transform(X_train)
svm = SVC()
# learn an SVM on the scaled training data
svm.fit(X_train_scaled, y_train)
# scale the test data and score the scaled data
X_test_scaled = scaler.transform(X_test)
print("Test score: {:.2f}".format(svm.score(X_test_scaled, y_test)))
Test score: 0.95
Parameter Selection with Preprocessing¶
- Scale이 적용된 훈련 데이터 전체를 GridSearchCV의 fit에 넣어줌
- GridSearchCV에서 자동으로 다시 훈련 데이터와 검증 데이터를 분리
- 검증 데이터는 Scale을 위한 fit에 활용되면 안되기 때문에 아래 코드는 문제가 있음
In [5]:
from sklearn.model_selection import GridSearchCV
# for illustration purposes only, don't use this code!
param_grid = {
'C': [0.001, 0.01, 0.1, 1, 10, 100],
'gamma': [0.001, 0.01, 0.1, 1, 10, 100]
}
grid = GridSearchCV(SVC(), param_grid=param_grid, cv=5)
grid.fit(X_train_scaled, y_train)
print("Best cross-validation accuracy: {:.2f}".format(grid.best_score_))
print("Best parameters: ", grid.best_params_)
X_test_scaled = scaler.transform(X_test)
print("Test set accuracy: {:.2f}".format(grid.score(X_test_scaled, y_test)))
Best cross-validation accuracy: 0.98
Best parameters: {'C': 1, 'gamma': 1}
Test set accuracy: 0.97
In [5]:
mglearn.plots.plot_improper_processing()
- 위 그림의 첫번째 경우의 문제점
- 검증 데이터의 정보가 Scale을 통한 데이터 변환시에 이미 노출이 됨.
Building Pipelines¶
- 교차 검증을 위한 훈련데이터와 검증데이터의 분할은 Scale과 같은 모든 데이터 변환 전처리 과정보다 앞서야 함.
- sklearn.pipeline.Pipeline
- Sequentially apply a list of transforms and a final estimator
- 교차 검증을 위한 훈련데이터/검증데이터 분할을 모든 데이터 변환 처리 보다 앞서게 할 수 있음.
In [9]:
from sklearn.pipeline import Pipeline
pipe = Pipeline([("scaler", MinMaxScaler()), ("svm", SVC())])
- 위와 같은 구성이 되면 pipe.steps에 Pipeline 정보가 들어감
- pipe.steps[0] <- ("scaler", MinMaxScaler())
- pipe.steps[0][1] <- MinMaxScaler()
- pipe.steps[1] <- ("svm", SVC())
- pipe.steps[1][1] <- SVC()
- pipe.steps[0] <- ("scaler", MinMaxScaler())
In [10]:
pipe.fit(X_train, y_train)
Out[10]:
Pipeline(steps=[('scaler', MinMaxScaler(copy=True, feature_range=(0, 1))), ('svm', SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape=None, degree=3, gamma='auto', kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False))])
In [11]:
print("Test score: {:.2f}".format(pipe.score(X_test, y_test)))
Test score: 0.95
Using Pipelines in Grid-searches¶
- 그리드 서치를 위한 후보 파라미터 설정시에 각 파라미터 이름 설정
- '단계의 이름' + '__' + '파라미터 이름'
In [12]:
param_grid = {
'svm__C': [0.001, 0.01, 0.1, 1, 10, 100],
'svm__gamma': [0.001, 0.01, 0.1, 1, 10, 100]
}
In [13]:
grid = GridSearchCV(pipe, param_grid=param_grid, cv=5)
grid.fit(X_train, y_train)
print("Best cross-validation accuracy: {:.2f}".format(grid.best_score_))
print("Test set score: {:.2f}".format(grid.score(X_test, y_test)))
print("Best parameters: {}".format(grid.best_params_))
Best cross-validation accuracy: 0.98
Test set score: 0.97
Best parameters: {'svm__C': 1, 'svm__gamma': 1}
In [11]:
mglearn.plots.plot_proper_processing()
[데이터 전처리 과정에 검증데이터 정보 누설에 따른 문제를 보여주는 예시]¶
- 아래에 임의로 생성한 데이터 X와 y는 전혀 연관관계가 없으므로 회귀 문제로 값을 예측하면 예측이 잘 안되는 것이 올바름
In [15]:
rnd = np.random.RandomState(seed=0)
X = rnd.normal(size=(100, 10000))
y = rnd.normal(size=(100,))
- Pipeline 사용없이 전체 데이터를 미리 전처리(변환)한 이후 그러한 데이터로 교차 검증을 하는 경우 예제 (잘못된 예제)
In [20]:
import warnings
warnings.filterwarnings('ignore')
from sklearn.feature_selection import SelectPercentile, f_regression
from sklearn.model_selection import cross_val_score
from sklearn.linear_model import Ridge
select = SelectPercentile(score_func=f_regression, percentile=5).fit(X, y)
X_selected = select.transform(X)
print("X_selected.shape: {}".format(X_selected.shape))
print("Cross-validation accuracy (cv only on ridge): {:.2f}".format(
np.mean(cross_val_score(Ridge(), X_selected, y, cv=5))
)
)
X_selected.shape: (100, 500) Cross-validation accuracy (cv only on ridge): 0.91
- Pipeline을 사용하여 전체 데이터의 전처리를 데이터 교차 검증과 내에서 수행하도록 하는 경우 예제 (올바른 예제)
In [21]:
select = SelectPercentile(score_func=f_regression, percentile=5)
model = Ridge()
pipe = Pipeline([("select", select), ("ridge", model)])
print("Cross-validation accuracy (pipeline): {:.2f}".format(
np.mean(cross_val_score(pipe, X, y, cv=5))
)
)
Cross-validation accuracy (pipeline): -0.25
The General Pipeline Interface¶
- Pipeline은 전처리나 분류에 국한하지 않고 어떠한 estimator와도 연결할 수 있음
- 예를 들어, 특성 추출, 특성 선택, 스케일 변경, 분류(또는 회귀, 군집)라는 총 4단계를 포함하는 파이프라인을 구성할 수 있음.
- Pipeline에 들어가는 estimator는 마지막 단계를 제외하고는 모두 fit과 transform을 지니고 있어야 함.
- 즉, 다음 단계를 위한 새로운 데이터 표현을 만들 수 있어야 함.
- Pipeline에 들어가는 마지막 estimator는 fit만 호출해도 됨.
In [16]:
def fit(self, X, y):
X_transformed = X
for name, estimator in self.steps[:-1]:
# iterate over all but the final step
# fit and transform the data
X_transformed = estimator.fit_transform(X_transformed, y)
# fit the last step
self.steps[-1][1].fit(X_transformed, y)
return self
In [17]:
def predict(self, X):
X_transformed = X
for step in self.steps[:-1]:
# iterate over all but the final step
# transform the data
X_transformed = step[1].transform(X_transformed)
# predict using the last step
return self.steps[-1][1].predict(X_transformed)
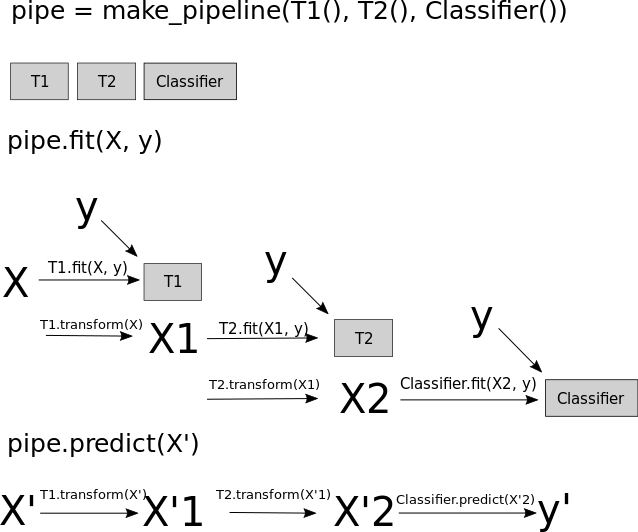
Convenient Pipeline creation with make_pipeline¶
In [22]:
from sklearn.pipeline import make_pipeline
# standard syntax
pipe_long = Pipeline([("scaler", MinMaxScaler()), ("svm", SVC(C=100))])
# abbreviated syntax
pipe_short = make_pipeline(MinMaxScaler(), SVC(C=100))
In [23]:
print("Pipeline steps:\n{}".format(pipe_short.steps))
Pipeline steps:
[('minmaxscaler', MinMaxScaler(copy=True, feature_range=(0, 1))), ('svc', SVC(C=100, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape=None, degree=3, gamma='auto', kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False))]
In [24]:
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
pipe = make_pipeline(StandardScaler(), PCA(n_components=2), StandardScaler())
print("Pipeline steps:\n{}".format(pipe.steps))
Pipeline steps:
[('standardscaler-1', StandardScaler(copy=True, with_mean=True, with_std=True)), ('pca', PCA(copy=True, iterated_power='auto', n_components=2, random_state=None,
svd_solver='auto', tol=0.0, whiten=False)), ('standardscaler-2', StandardScaler(copy=True, with_mean=True, with_std=True))]
Accessing step attributes¶
In [25]:
# fit the pipeline defined before to the cancer dataset
print("cancer data shape: {}".format(cancer.data.shape))
pipe.fit(cancer.data)
# extract the first two principal components from the "pca" step
components = pipe.named_steps["pca"].components_
print("components.shape: {}".format(components.shape))
cancer data shape: (569, 30) components.shape: (2, 30)
Accessing Attributes in a Pipeline inside GridSearchCV¶
In [26]:
from sklearn.linear_model import LogisticRegression
pipe = make_pipeline(StandardScaler(), LogisticRegression())
In [27]:
param_grid = {
'logisticregression__C': [0.01, 0.1, 1, 10, 100]
}
In [28]:
X_train, X_test, y_train, y_test = train_test_split(cancer.data, cancer.target, random_state=4)
grid = GridSearchCV(pipe, param_grid, cv=5)
grid.fit(X_train, y_train)
Out[28]:
GridSearchCV(cv=5, error_score='raise',
estimator=Pipeline(steps=[('standardscaler', StandardScaler(copy=True, with_mean=True, with_std=True)), ('logisticregression', LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, max_iter=100, multi_class='ovr', n_jobs=1,
penalty='l2', random_state=None, solver='liblinear', tol=0.0001,
verbose=0, warm_start=False))]),
fit_params={}, iid=True, n_jobs=1,
param_grid={'logisticregression__C': [0.01, 0.1, 1, 10, 100]},
pre_dispatch='2*n_jobs', refit=True, return_train_score=True,
scoring=None, verbose=0)
In [29]:
print("Best estimator:\n{}".format(grid.best_estimator_))
Best estimator:
Pipeline(steps=[('standardscaler', StandardScaler(copy=True, with_mean=True, with_std=True)), ('logisticregression', LogisticRegression(C=0.1, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, max_iter=100, multi_class='ovr', n_jobs=1,
penalty='l2', random_state=None, solver='liblinear', tol=0.0001,
verbose=0, warm_start=False))])
In [26]:
print("Logistic regression step:\n{}".format(
grid.best_estimator_.named_steps["logisticregression"]))
Logistic regression step:
LogisticRegression(C=0.1, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, max_iter=100, multi_class='ovr', n_jobs=1,
penalty='l2', random_state=None, solver='liblinear', tol=0.0001,
verbose=0, warm_start=False)
In [30]:
print("Logistic regression coefficients:\n{}".format(
grid.best_estimator_.named_steps["logisticregression"].coef_
)
)
Logistic regression coefficients: [[-0.389 -0.375 -0.376 -0.396 -0.115 0.017 -0.355 -0.39 -0.058 0.209 -0.495 -0.004 -0.371 -0.383 -0.045 0.198 0.004 -0.049 0.21 0.224 -0.547 -0.525 -0.499 -0.515 -0.393 -0.123 -0.388 -0.417 -0.325 -0.139]]
Grid-searching preprocessing steps and model parameters¶
In [33]:
from sklearn.datasets import load_boston
from sklearn.preprocessing import PolynomialFeatures
boston = load_boston()
X_train, X_test, y_train, y_test = train_test_split(boston.data, boston.target, random_state=0)
pipe = make_pipeline(
StandardScaler(),
PolynomialFeatures(),
Ridge()
)
In [34]:
param_grid = {
'polynomialfeatures__degree': [1, 2, 3],
'ridge__alpha': [0.001, 0.01, 0.1, 1, 10, 100]
}
In [ ]:
grid = GridSearchCV(pipe, param_grid=param_grid, cv=5, n_jobs=-1)
grid.fit(X_train, y_train)
In [31]:
mglearn.tools.heatmap(
grid.cv_results_['mean_test_score'].reshape(3, -1),
xlabel="ridge__alpha",
ylabel="polynomialfeatures__degree",
xticklabels=param_grid['ridge__alpha'],
yticklabels=param_grid['polynomialfeatures__degree'], vmin=0
)
Out[31]:
<matplotlib.collections.PolyCollection at 0x7fecac083ba8>
In [32]:
print("Best parameters: {}".format(grid.best_params_))
Best parameters: {'polynomialfeatures__degree': 2, 'ridge__alpha': 10}
In [33]:
print("Test-set score: {:.2f}".format(grid.score(X_test, y_test)))
Test-set score: 0.77
In [34]:
param_grid = {
'ridge__alpha': [0.001, 0.01, 0.1, 1, 10, 100]
}
pipe = make_pipeline(StandardScaler(), Ridge())
grid = GridSearchCV(pipe, param_grid, cv=5)
grid.fit(X_train, y_train)
print("Score without poly features: {:.2f}".format(grid.score(X_test, y_test)))
Score without poly features: 0.63
In [35]:
pipe = Pipeline([('preprocessing', StandardScaler()), ('classifier', SVC())])
In [36]:
from sklearn.ensemble import RandomForestClassifier
param_grid = [
{'classifier': [SVC()],
'preprocessing': [StandardScaler(), None],
'classifier__gamma': [0.001, 0.01, 0.1, 1, 10, 100],
'classifier__C': [0.001, 0.01, 0.1, 1, 10, 100]
},
{'classifier': [RandomForestClassifier(n_estimators=100)],
'preprocessing': [None],
'classifier__max_features': [1, 2, 3]
}
]
In [37]:
X_train, X_test, y_train, y_test = train_test_split(cancer.data, cancer.target, random_state=0)
grid = GridSearchCV(pipe, param_grid, cv=5)
grid.fit(X_train, y_train)
print("Best params:\n{}\n".format(grid.best_params_))
print("Best cross-validation score: {:.2f}".format(grid.best_score_))
print("Test-set score: {:.2f}".format(grid.score(X_test, y_test)))
Best params:
{'classifier': SVC(C=10, cache_size=200, class_weight=None, coef0=0.0,
decision_function_shape='ovr', degree=3, gamma=0.01, kernel='rbf',
max_iter=-1, probability=False, random_state=None, shrinking=True,
tol=0.001, verbose=False), 'classifier__C': 10, 'classifier__gamma': 0.01, 'preprocessing': StandardScaler(copy=True, with_mean=True, with_std=True)}
Best cross-validation score: 0.99
Test-set score: 0.98