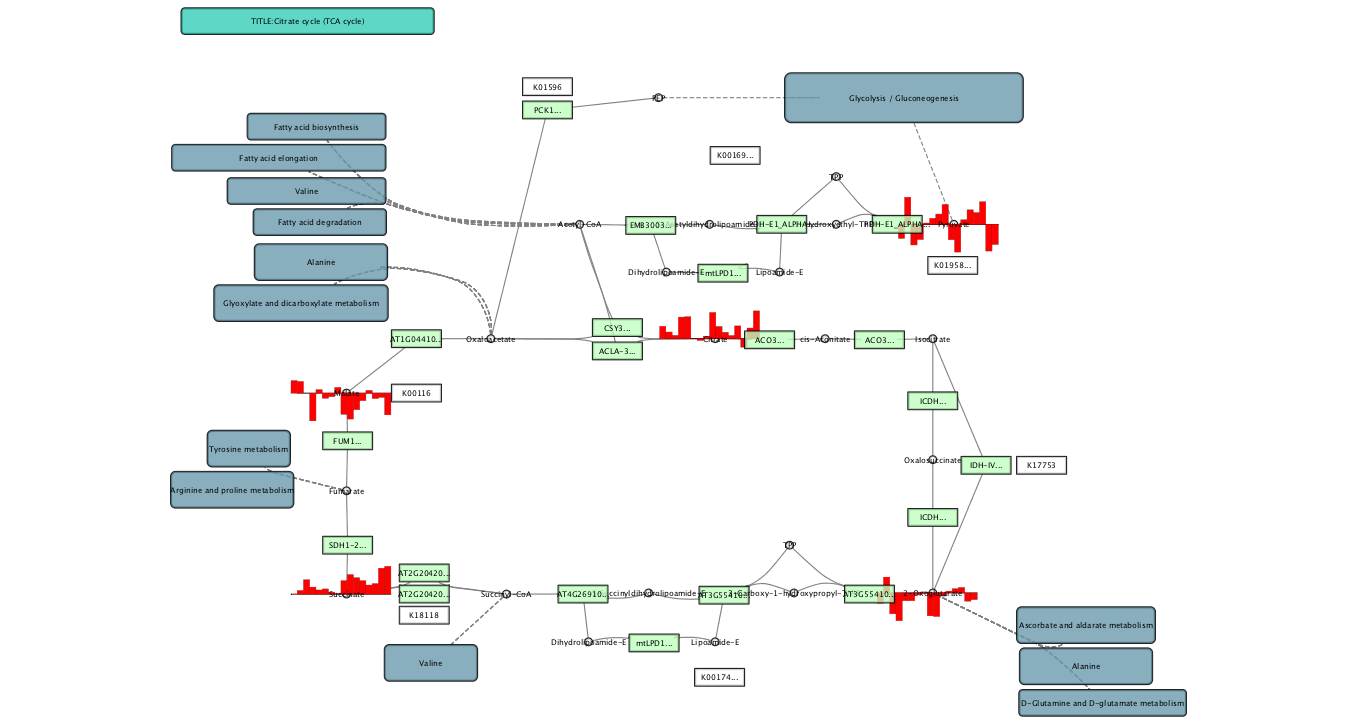
Visualizing time series metabolome profile¶
by Kozo Nishida (Riken, Japan)
Software Requirments¶
Please install the following software packages to run this workflow:
Background¶
This is a sample workflow to automate complex Cytoscape data integaration/visualization process. Please read the following document for more background:
In [10]:
import json
import requests
import pandas as pd
PORT_NUMBER = 1234
BASE_URL = "http://localhost:" + str(PORT_NUMBER) + "/v1/"
HEADERS = {'Content-Type': 'application/json'}
Load a KGML pathway data file from KEGG REST API¶
In [11]:
pathway_location = "http://rest.kegg.jp/get/ath00020/kgml"
res1 = requests.post(BASE_URL + "networks?source=url", data=json.dumps([pathway_location]), headers=HEADERS)
result = json.loads(res1.content)
pathway_suid = result[0]["networkSUID"][0]
print("Pathway SUID = " + str(pathway_suid))
Pathway SUID = 71718
Load table data file as Pandas DataFrame¶
In [12]:
profile_csv = "https://raw.githubusercontent.com/idekerlab/KEGGscape/develop/wiki/data/light-dark-20.csv"
profile_df = pd.read_csv(profile_csv)
profile_df.head()
Out[12]:
| Metabolite | KEGG | ld20t14 | ld20t16 | ld20t20 | ld20t24 | ld20t28 | ld20t32 | ld20t36 | ld20t40 | ld20t44 | ld20t48 | ld20t52 | ld20t56 | ld20t60 | ld20t64 | ld20t68 | ld20t72 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Alanine | C00041 | -0.051007 | 0.040631 | -0.165335 | -0.126382 | -0.093879 | -0.102625 | 0.009509 | 0.051824 | -0.137530 | -0.140400 | -0.116827 | -0.060302 | 0.027001 | 0.081365 | -0.060153 | -0.097988 |
| 1 | Valine | C00183 | -0.025403 | -0.003585 | -0.058986 | -0.069280 | -0.129784 | -0.092210 | 0.009129 | -0.068003 | -0.105905 | -0.097984 | -0.095181 | -0.021911 | -0.021224 | -0.028314 | -0.032834 | -0.025435 |
| 2 | Leucine | C00123 | -0.005918 | 0.058287 | -0.049900 | -0.075853 | -0.098339 | -0.003847 | 0.056037 | -0.104399 | -0.122298 | -0.098051 | -0.082529 | 0.027760 | -0.006289 | -0.048610 | -0.042620 | -0.000897 |
| 3 | Isoleucine | C00407 | 0.079870 | 0.101324 | -0.103993 | -0.041534 | -0.020148 | 0.066486 | 0.091006 | -0.027547 | -0.193494 | -0.144676 | -0.015375 | 0.070540 | 0.118164 | 0.013795 | -0.060418 | -0.042314 |
| 4 | Glycine | C00037 | -0.056155 | -0.185757 | -0.857195 | -0.725952 | -0.051028 | 0.069038 | 0.042867 | 0.098185 | -0.445921 | -0.893788 | -0.201299 | 0.000000 | -0.064322 | -0.090597 | -0.637518 | -0.874438 |
Convert the DataFrame to JSON and send it to Cytoscape¶
In [13]:
profile = json.loads(profile_df.to_json(orient="records"))
# print(json.dumps(profile, indent=4))
In [14]:
new_table_data = {
"key": "KEGG_NODE_LABEL",
"dataKey": "KEGG",
"data": profile
}
update_table_url = BASE_URL + "networks/" + str(pathway_suid) + "/tables/defaultnode"
requests.put(update_table_url, data=json.dumps(new_table_data), headers=HEADERS)
Out[14]:
<Response [200]>
Set values to the chart column¶
In [15]:
chart_entry = 'barchart: attributelist="ld20t14,ld20t16,ld20t20,ld20t24,ld20t28,ld20t32,ld20t36,ld20t40,ld20t44,ld20t48,ld20t52,ld20t56,ld20t60,ld20t64,ld20t68,ld20t72" colorlist="up:red,zero:red,down:red" showlabels="false"'
target_row_url = BASE_URL + "networks/" + str(pathway_suid) + "/tables/defaultnode/columns/KEGG"
res2 = requests.get(target_row_url)
matched = json.loads(res2.content)["values"]
df2 = pd.DataFrame(columns=["id", "chart"]);
df2["id"] = matched
df2["chart"] = chart_entry
data = json.loads(df2.to_json(orient="records"))
chart_data = {
"key": "KEGG",
"dataKey": "id",
"data": data
}
requests.put(update_table_url, data=json.dumps(chart_data), headers=HEADERS)
Out[15]:
<Response [200]>
Create Visual Style for Custom Mapping¶
In [16]:
custom_graphics_mapping = {
"mappingType" : "passthrough",
"mappingColumn" : "chart",
"mappingColumnType" : "String",
"visualProperty" : "NODE_CUSTOMGRAPHICS_1"
}
style_url = BASE_URL + "styles/KEGG Style/mappings"
requests.post(style_url, data=json.dumps([custom_graphics_mapping]), headers=HEADERS)
Out[16]:
<Response [201]>
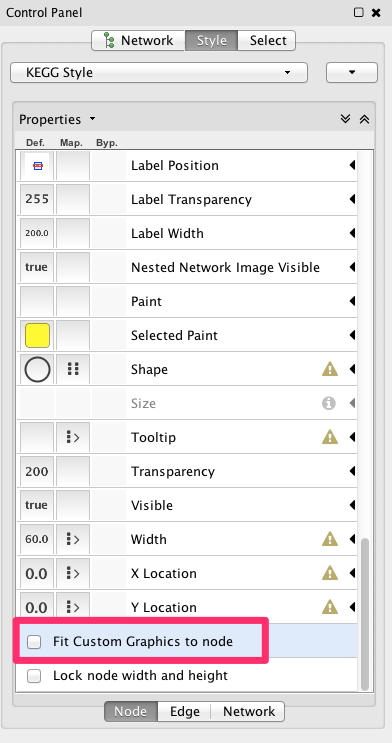
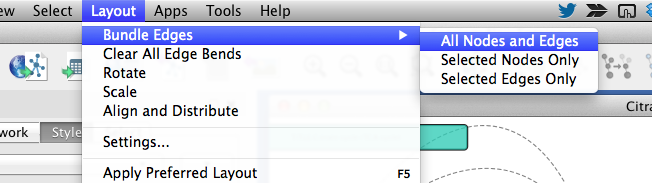
Uncheck the Fit Custom Graphics to node box, and bundle edges from menubar.
You will see