Mushroom Dataset¶
1. Data Set Desciption¶
- http://archive.ics.uci.edu/ml/datasets/Mushroom
- Attribute Information
- 0 - classes (target attribute): edible=e, poisonous=p
- 1 - cap-shape: bell=b, conical=c, convex=x, flat=f, knobbed=k, sunken=s
- 2 - cap-surface: fibrous=f, grooves=g, scaly=y, smooth=s
- 3 - cap-color: brown=n, buff=b, cinnamon=c, gray=g, green=r, pink=p, purple=u, red=e, white=w, yellow=y
- 4 - bruises: bruises=t, no=f
- 5 - odor: almond=a, anise=l, creosote=c, fishy=y, foul=f, musty=m, none=n, pungent=p, spicy=s
- 6 - gill-attachment: attached=a, descending=d, free=f, notched=n
- 7 - gill-spacing: close=c,crowded=w,distant=d
- 8 - gill-size: broad=b, narrow=n
- 9 - gill-color: black=k, brown=n, buff=b, chocolate=h, gray=g, green=r, orange=o, pink=p, purple=u, red=e, white=w, yellow=y
- 10 - stalk-shape: enlarging=e, tapering=t
- 11 - stalk-root: bulbous=b, club=c, cup=u, equal=e, rhizomorphs=z, rooted=r, missing=?
- 12 - stalk-surface-above-ring: fibrous=f, scaly=y, silky=k, smooth=s
- 13 - stalk-surface-below-ring: fibrous=f, scaly=y, silky=k, smooth=s
- 14 - stalk-color-above-ring: brown=n, buff=b, cinnamon=c, gray=g, orange=o, pink=p, red=e, white=w, yellow=y
- 15 - stalk-color-below-ring: brown=n, buff=b, cinnamon=c, gray=g, orange=o, pink=p, red=e, white=w, yellow=y
- 16 - veil-type: partial=p, universal=u
- 17 - veil-color: brown=n, orange=o, white=w, yellow=y
- 18 - ring-number: none=n, one=o, two=t
- 19 - ring-type: cobwebby=c, evanescent=e, flaring=f, large=l, none=n, pendant=p, sheathing=s, zone=z
- 20 - spore-print-color: black=k, brown=n, buff=b, chocolate=h, green=r, orange=o, purple=u, white=w, yellow=y
- 21 - population: abundant=a, clustered=c, numerous=n, scattered=s, several=v, solitary=y
- 22 - habitat: grasses=g, leaves=l, meadows=m, paths=p, urban=u, waste=w, woods=d
2. Pandas DataFame 다루기¶
1) Loading Data¶
In [1]:
import urllib2
from scipy import stats
from pandas import Series, DataFrame
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
%matplotlib inline
path = 'http://archive.ics.uci.edu/ml/machine-learning-databases/mushroom/agaricus-lepiota.data'
raw_csv = urllib2.urlopen(path)
col_names = range(23)
df = pd.read_csv(raw_csv, names = col_names)
In [2]:
df.head()
Out[2]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | p | x | s | n | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | k | s | u |
| 1 | e | x | s | y | t | a | f | c | b | k | ... | s | w | w | p | w | o | p | n | n | g |
| 2 | e | b | s | w | t | l | f | c | b | n | ... | s | w | w | p | w | o | p | n | n | m |
| 3 | p | x | y | w | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | k | s | u |
| 4 | e | x | s | g | f | n | f | w | b | k | ... | s | w | w | p | w | o | e | n | a | g |
5 rows × 23 columns
In [3]:
df.describe()
Out[3]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | ... | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 | 8124 |
| unique | 2 | 6 | 4 | 10 | 2 | 9 | 2 | 2 | 2 | 12 | ... | 4 | 9 | 9 | 1 | 4 | 3 | 5 | 9 | 6 | 7 |
| top | e | x | y | n | f | n | f | c | b | b | ... | s | w | w | p | w | o | p | w | v | d |
| freq | 4208 | 3656 | 3244 | 2284 | 4748 | 3528 | 7914 | 6812 | 5612 | 1728 | ... | 4936 | 4464 | 4384 | 8124 | 7924 | 7488 | 3968 | 2388 | 4040 | 3148 |
4 rows × 23 columns
2) Missing Value 찾기¶
In [4]:
df.isnull().head()
Out[4]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | False | False | False | False | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| 1 | False | False | False | False | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| 2 | False | False | False | False | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| 3 | False | False | False | False | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
| 4 | False | False | False | False | False | False | False | False | False | False | ... | False | False | False | False | False | False | False | False | False | False |
5 rows × 23 columns
In [5]:
df.isnull().values
Out[5]:
array([[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
...,
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False],
[False, False, False, ..., False, False, False]], dtype=bool)
In [6]:
df.isnull().values.any()
Out[6]:
False
2) DataFrame의 groupby 학습¶
In [7]:
temp_df = DataFrame([[1, 2], [1, 4], [5, 6], [5, 4], [5, 4]], columns=['A', 'B'])
In [8]:
temp_df
Out[8]:
| A | B | |
|---|---|---|
| 0 | 1 | 2 |
| 1 | 1 | 4 |
| 2 | 5 | 6 |
| 3 | 5 | 4 |
| 4 | 5 | 4 |
- 컬럼 A로 그룹핑¶
In [9]:
groupby_temp_df = temp_df.groupby('A')
In [10]:
groupby_temp_df.groups
Out[10]:
{1: [0, 1], 5: [2, 3, 4]}
In [11]:
groupby_temp_df.get_group(1)
Out[11]:
| A | B | |
|---|---|---|
| 0 | 1 | 2 |
| 1 | 1 | 4 |
In [12]:
groupby_temp_df.get_group(5)
Out[12]:
| A | B | |
|---|---|---|
| 2 | 5 | 6 |
| 3 | 5 | 4 |
| 4 | 5 | 4 |
- 각 그룹마다 존재하는 아이템의 개수 반환
In [13]:
groupby_temp_df.count()
Out[13]:
| B | |
|---|---|
| A | |
| 1 | 2 |
| 5 | 3 |
- 각 그룹마다 n 개씩의 row를 반환하는 방법
In [14]:
groupby_temp_df.head(1)
Out[14]:
| A | B | |
|---|---|---|
| 0 | 1 | 2 |
| 2 | 5 | 6 |
In [15]:
groupby_temp_df.describe()
Out[15]:
| A | B | ||
|---|---|---|---|
| A | |||
| 1 | count | 2.0 | 2.000000 |
| mean | 1.0 | 3.000000 | |
| std | 0.0 | 1.414214 | |
| min | 1.0 | 2.000000 | |
| 25% | 1.0 | 2.500000 | |
| 50% | 1.0 | 3.000000 | |
| 75% | 1.0 | 3.500000 | |
| max | 1.0 | 4.000000 | |
| 5 | count | 3.0 | 3.000000 |
| mean | 5.0 | 4.666667 | |
| std | 0.0 | 1.154701 | |
| min | 5.0 | 4.000000 | |
| 25% | 5.0 | 4.000000 | |
| 50% | 5.0 | 4.000000 | |
| 75% | 5.0 | 5.000000 | |
| max | 5.0 | 6.000000 |
- 컬럼 B로 그룹핑¶
In [16]:
groupby_temp_df2 = temp_df.groupby('B')
In [17]:
groupby_temp_df2.groups
Out[17]:
{2: [0], 4: [1, 3, 4], 6: [2]}
In [18]:
groupby_temp_df2.get_group(2)
Out[18]:
| A | B | |
|---|---|---|
| 0 | 1 | 2 |
In [19]:
groupby_temp_df2.get_group(4)
Out[19]:
| A | B | |
|---|---|---|
| 1 | 1 | 4 |
| 3 | 5 | 4 |
| 4 | 5 | 4 |
In [20]:
groupby_temp_df2.get_group(6)
Out[20]:
| A | B | |
|---|---|---|
| 2 | 5 | 6 |
In [21]:
groupby_temp_df2.count()
Out[21]:
| A | |
|---|---|
| B | |
| 2 | 1 |
| 4 | 3 |
| 6 | 1 |
In [22]:
groupby_temp_df2.head(1)
Out[22]:
| A | B | |
|---|---|---|
| 0 | 1 | 2 |
| 1 | 1 | 4 |
| 2 | 5 | 6 |
In [23]:
groupby_temp_df2.describe()
Out[23]:
| A | B | ||
|---|---|---|---|
| B | |||
| 2 | count | 1.000000 | 1.0 |
| mean | 1.000000 | 2.0 | |
| std | NaN | NaN | |
| min | 1.000000 | 2.0 | |
| 25% | 1.000000 | 2.0 | |
| 50% | 1.000000 | 2.0 | |
| 75% | 1.000000 | 2.0 | |
| max | 1.000000 | 2.0 | |
| 4 | count | 3.000000 | 3.0 |
| mean | 3.666667 | 4.0 | |
| std | 2.309401 | 0.0 | |
| min | 1.000000 | 4.0 | |
| 25% | 3.000000 | 4.0 | |
| 50% | 5.000000 | 4.0 | |
| 75% | 5.000000 | 4.0 | |
| max | 5.000000 | 4.0 | |
| 6 | count | 1.000000 | 1.0 |
| mean | 5.000000 | 6.0 | |
| std | NaN | NaN | |
| min | 5.000000 | 6.0 | |
| 25% | 5.000000 | 6.0 | |
| 50% | 5.000000 | 6.0 | |
| 75% | 5.000000 | 6.0 | |
| max | 5.000000 | 6.0 |
4) Mushroom 데이터의 각 속성별로 그룹핑 작업을 위한 정보 확인¶
In [24]:
df[[0,1]].head(5)
Out[24]:
| 0 | 1 | |
|---|---|---|
| 0 | p | x |
| 1 | e | x |
| 2 | e | b |
| 3 | p | x |
| 4 | e | x |
In [25]:
a = df[[0,1]].groupby(1)
In [26]:
print type(a)
<class 'pandas.core.groupby.DataFrameGroupBy'>
In [27]:
a.count()
Out[27]:
| 0 | |
|---|---|
| 1 | |
| b | 452 |
| c | 4 |
| f | 3152 |
| k | 828 |
| s | 32 |
| x | 3656 |
In [28]:
a.head(2)
Out[28]:
| 0 | 1 | |
|---|---|---|
| 0 | p | x |
| 1 | e | x |
| 2 | e | b |
| 6 | e | b |
| 15 | e | s |
| 16 | e | f |
| 25 | p | f |
| 36 | e | s |
| 4276 | e | k |
| 4291 | e | k |
| 5126 | p | c |
| 5508 | p | c |
In [29]:
a.groups.keys()
Out[29]:
['c', 'b', 'f', 'k', 's', 'x']
In [30]:
a.size()
Out[30]:
1 b 452 c 4 f 3152 k 828 s 32 x 3656 dtype: int64
In [31]:
a.ngroups
Out[31]:
6
In [32]:
a.get_group("c")
Out[32]:
| 0 | 1 | |
|---|---|---|
| 5126 | p | c |
| 5508 | p | c |
| 7401 | p | c |
| 7706 | p | c |
In [33]:
a.describe()
Out[33]:
| 0 | 1 | ||
|---|---|---|---|
| 1 | |||
| b | count | 452 | 452 |
| unique | 2 | 1 | |
| top | e | b | |
| freq | 404 | 452 | |
| c | count | 4 | 4 |
| unique | 1 | 1 | |
| top | p | c | |
| freq | 4 | 4 | |
| f | count | 3152 | 3152 |
| unique | 2 | 1 | |
| top | e | f | |
| freq | 1596 | 3152 | |
| k | count | 828 | 828 |
| unique | 2 | 1 | |
| top | p | k | |
| freq | 600 | 828 | |
| s | count | 32 | 32 |
| unique | 1 | 1 | |
| top | e | s | |
| freq | 32 | 32 | |
| x | count | 3656 | 3656 |
| unique | 2 | 1 | |
| top | e | x | |
| freq | 1948 | 3656 |
5) 각 속성별 주요 정보를 담은 df_per_attr 사전 만들기¶
In [34]:
df_per_attr = {}
for i in range(1, 23):
df_per_attr[i] = {}
groupby_df = df[[0, i]].groupby(i)
df_per_attr[i]['ngorups'] = groupby_df.ngroups
df_per_attr[i]['group_keys'] = groupby_df.groups.keys()
df_per_attr[i]['subgroups'] = {}
for j in range(groupby_df.ngroups):
df_per_attr[i]['subgroups'][j] = groupby_df.get_group(df_per_attr[i]['group_keys'][j])
In [35]:
df_per_attr[1]['group_keys']
Out[35]:
['c', 'b', 'f', 'k', 's', 'x']
In [36]:
df_per_attr[1]['subgroups'][0]
Out[36]:
| 0 | 1 | |
|---|---|---|
| 5126 | p | c |
| 5508 | p | c |
| 7401 | p | c |
| 7706 | p | c |
6) Categorical Attribute를 Numerical Attribute로 변환¶
In [37]:
df[0] = df[0].map({'p': 1, 'e': 0})
df
Out[37]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | x | s | n | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | k | s | u |
| 1 | 0 | x | s | y | t | a | f | c | b | k | ... | s | w | w | p | w | o | p | n | n | g |
| 2 | 0 | b | s | w | t | l | f | c | b | n | ... | s | w | w | p | w | o | p | n | n | m |
| 3 | 1 | x | y | w | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | k | s | u |
| 4 | 0 | x | s | g | f | n | f | w | b | k | ... | s | w | w | p | w | o | e | n | a | g |
| 5 | 0 | x | y | y | t | a | f | c | b | n | ... | s | w | w | p | w | o | p | k | n | g |
| 6 | 0 | b | s | w | t | a | f | c | b | g | ... | s | w | w | p | w | o | p | k | n | m |
| 7 | 0 | b | y | w | t | l | f | c | b | n | ... | s | w | w | p | w | o | p | n | s | m |
| 8 | 1 | x | y | w | t | p | f | c | n | p | ... | s | w | w | p | w | o | p | k | v | g |
| 9 | 0 | b | s | y | t | a | f | c | b | g | ... | s | w | w | p | w | o | p | k | s | m |
| 10 | 0 | x | y | y | t | l | f | c | b | g | ... | s | w | w | p | w | o | p | n | n | g |
| 11 | 0 | x | y | y | t | a | f | c | b | n | ... | s | w | w | p | w | o | p | k | s | m |
| 12 | 0 | b | s | y | t | a | f | c | b | w | ... | s | w | w | p | w | o | p | n | s | g |
| 13 | 1 | x | y | w | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | n | v | u |
| 14 | 0 | x | f | n | f | n | f | w | b | n | ... | f | w | w | p | w | o | e | k | a | g |
| 15 | 0 | s | f | g | f | n | f | c | n | k | ... | s | w | w | p | w | o | p | n | y | u |
| 16 | 0 | f | f | w | f | n | f | w | b | k | ... | s | w | w | p | w | o | e | n | a | g |
| 17 | 1 | x | s | n | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | k | s | g |
| 18 | 1 | x | y | w | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | n | s | u |
| 19 | 1 | x | s | n | t | p | f | c | n | k | ... | s | w | w | p | w | o | p | n | s | u |
| 20 | 0 | b | s | y | t | a | f | c | b | k | ... | s | w | w | p | w | o | p | n | s | m |
| 21 | 1 | x | y | n | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | n | v | g |
| 22 | 0 | b | y | y | t | l | f | c | b | k | ... | s | w | w | p | w | o | p | n | s | m |
| 23 | 0 | b | y | w | t | a | f | c | b | w | ... | s | w | w | p | w | o | p | n | n | m |
| 24 | 0 | b | s | w | t | l | f | c | b | g | ... | s | w | w | p | w | o | p | k | s | m |
| 25 | 1 | f | s | w | t | p | f | c | n | n | ... | s | w | w | p | w | o | p | n | v | g |
| 26 | 0 | x | y | y | t | a | f | c | b | n | ... | s | w | w | p | w | o | p | n | n | m |
| 27 | 0 | x | y | w | t | l | f | c | b | w | ... | s | w | w | p | w | o | p | n | n | m |
| 28 | 0 | f | f | n | f | n | f | c | n | k | ... | s | w | w | p | w | o | p | k | y | u |
| 29 | 0 | x | s | y | t | a | f | w | n | n | ... | s | w | w | p | w | o | p | n | v | d |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 8094 | 0 | b | s | g | f | n | f | w | b | g | ... | s | w | w | p | w | t | p | w | n | g |
| 8095 | 1 | x | y | c | f | m | f | c | b | y | ... | y | c | c | p | w | n | n | w | c | d |
| 8096 | 0 | k | f | w | f | n | f | w | b | w | ... | s | w | w | p | w | t | p | w | n | g |
| 8097 | 1 | k | y | n | f | s | f | c | n | b | ... | k | p | p | p | w | o | e | w | v | l |
| 8098 | 1 | k | s | e | f | y | f | c | n | b | ... | k | w | p | p | w | o | e | w | v | d |
| 8099 | 0 | k | f | w | f | n | f | w | b | w | ... | k | w | w | p | w | t | p | w | s | g |
| 8100 | 0 | f | s | n | f | n | a | c | b | o | ... | s | o | o | p | n | o | p | b | v | l |
| 8101 | 1 | k | s | e | f | s | f | c | n | b | ... | s | p | w | p | w | o | e | w | v | p |
| 8102 | 0 | x | s | n | f | n | a | c | b | y | ... | s | o | o | p | n | o | p | n | c | l |
| 8103 | 0 | k | s | n | f | n | a | c | b | y | ... | s | o | o | p | n | o | p | o | c | l |
| 8104 | 0 | k | s | n | f | n | a | c | b | y | ... | s | o | o | p | o | o | p | n | v | l |
| 8105 | 0 | k | s | n | f | n | a | c | b | y | ... | s | o | o | p | n | o | p | y | v | l |
| 8106 | 0 | k | s | n | f | n | a | c | b | o | ... | s | o | o | p | o | o | p | n | v | l |
| 8107 | 0 | x | s | n | f | n | a | c | b | y | ... | s | o | o | p | o | o | p | n | c | l |
| 8108 | 1 | k | y | e | f | y | f | c | n | b | ... | s | p | w | p | w | o | e | w | v | l |
| 8109 | 0 | b | s | w | f | n | f | w | b | w | ... | s | w | w | p | w | t | p | w | n | g |
| 8110 | 0 | x | s | n | f | n | a | c | b | o | ... | s | o | o | p | o | o | p | n | v | l |
| 8111 | 0 | k | s | w | f | n | f | w | b | p | ... | s | w | w | p | w | t | p | w | n | g |
| 8112 | 0 | k | s | n | f | n | a | c | b | o | ... | s | o | o | p | n | o | p | b | v | l |
| 8113 | 1 | k | y | e | f | y | f | c | n | b | ... | k | p | p | p | w | o | e | w | v | d |
| 8114 | 1 | f | y | c | f | m | a | c | b | y | ... | y | c | c | p | w | n | n | w | c | d |
| 8115 | 0 | x | s | n | f | n | a | c | b | y | ... | s | o | o | p | o | o | p | o | v | l |
| 8116 | 1 | k | y | n | f | s | f | c | n | b | ... | k | p | w | p | w | o | e | w | v | l |
| 8117 | 1 | k | s | e | f | y | f | c | n | b | ... | s | p | w | p | w | o | e | w | v | d |
| 8118 | 1 | k | y | n | f | f | f | c | n | b | ... | s | p | w | p | w | o | e | w | v | d |
| 8119 | 0 | k | s | n | f | n | a | c | b | y | ... | s | o | o | p | o | o | p | b | c | l |
| 8120 | 0 | x | s | n | f | n | a | c | b | y | ... | s | o | o | p | n | o | p | b | v | l |
| 8121 | 0 | f | s | n | f | n | a | c | b | n | ... | s | o | o | p | o | o | p | b | c | l |
| 8122 | 1 | k | y | n | f | y | f | c | n | b | ... | k | w | w | p | w | o | e | w | v | l |
| 8123 | 0 | x | s | n | f | n | a | c | b | y | ... | s | o | o | p | o | o | p | o | c | l |
8124 rows × 23 columns
In [38]:
print df.shape[0], df.shape[1]
8124 23
In [39]:
num_columns = df.shape[1]
map_dic = {}
for i in range(num_columns):
unique_array = df[i].unique()
N = len(unique_array)
map_dic[i] = {}
for j in range(N):
map_dic[i][unique_array[j]] = j
df[i] = df[i].map(map_dic[i])
df
Out[39]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 1 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| 2 | 1 | 1 | 0 | 2 | 0 | 2 | 0 | 0 | 1 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| 3 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| 4 | 1 | 0 | 0 | 3 | 1 | 3 | 0 | 1 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 1 |
| 5 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 |
| 6 | 1 | 1 | 0 | 2 | 0 | 1 | 0 | 0 | 1 | 2 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 |
| 7 | 1 | 1 | 1 | 2 | 0 | 2 | 0 | 0 | 1 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| 8 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 3 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 |
| 9 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 2 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 10 | 1 | 0 | 1 | 1 | 0 | 2 | 0 | 0 | 1 | 2 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 1 |
| 11 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 12 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 4 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 |
| 13 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 0 |
| 14 | 1 | 0 | 2 | 0 | 1 | 3 | 0 | 1 | 1 | 1 | ... | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 | 1 |
| 15 | 1 | 2 | 2 | 3 | 1 | 3 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 0 |
| 16 | 1 | 3 | 2 | 2 | 1 | 3 | 0 | 1 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 1 |
| 17 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| 18 | 0 | 0 | 1 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 19 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| 20 | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| 21 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 1 |
| 22 | 1 | 1 | 1 | 1 | 0 | 2 | 0 | 0 | 1 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| 23 | 1 | 1 | 1 | 2 | 0 | 1 | 0 | 0 | 1 | 4 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| 24 | 1 | 1 | 0 | 2 | 0 | 2 | 0 | 0 | 1 | 2 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 |
| 25 | 0 | 3 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 1 |
| 26 | 1 | 0 | 1 | 1 | 0 | 1 | 0 | 0 | 1 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| 27 | 1 | 0 | 1 | 2 | 0 | 2 | 0 | 0 | 1 | 4 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 2 |
| 28 | 1 | 3 | 2 | 0 | 1 | 3 | 0 | 0 | 0 | 0 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 |
| 29 | 1 | 0 | 0 | 1 | 0 | 1 | 0 | 1 | 0 | 1 | ... | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 | 3 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 8094 | 1 | 1 | 0 | 3 | 1 | 3 | 0 | 1 | 1 | 2 | ... | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 1 | 1 |
| 8095 | 0 | 0 | 1 | 8 | 1 | 8 | 0 | 0 | 1 | 10 | ... | 2 | 7 | 8 | 0 | 0 | 2 | 4 | 4 | 5 | 3 |
| 8096 | 1 | 4 | 2 | 2 | 1 | 3 | 0 | 1 | 1 | 4 | ... | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 1 | 1 |
| 8097 | 0 | 4 | 1 | 0 | 1 | 7 | 0 | 0 | 0 | 8 | ... | 3 | 2 | 1 | 0 | 0 | 0 | 1 | 4 | 3 | 6 |
| 8098 | 0 | 4 | 0 | 4 | 1 | 6 | 0 | 0 | 0 | 8 | ... | 3 | 0 | 1 | 0 | 0 | 0 | 1 | 4 | 3 | 3 |
| 8099 | 1 | 4 | 2 | 2 | 1 | 3 | 0 | 1 | 1 | 4 | ... | 3 | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 0 | 1 |
| 8100 | 1 | 3 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 11 | ... | 0 | 6 | 7 | 0 | 1 | 0 | 0 | 8 | 3 | 6 |
| 8101 | 0 | 4 | 0 | 4 | 1 | 7 | 0 | 0 | 0 | 8 | ... | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 4 | 3 | 4 |
| 8102 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 10 | ... | 0 | 6 | 7 | 0 | 1 | 0 | 0 | 1 | 5 | 6 |
| 8103 | 1 | 4 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 10 | ... | 0 | 6 | 7 | 0 | 1 | 0 | 0 | 6 | 5 | 6 |
| 8104 | 1 | 4 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 10 | ... | 0 | 6 | 7 | 0 | 2 | 0 | 0 | 1 | 3 | 6 |
| 8105 | 1 | 4 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 10 | ... | 0 | 6 | 7 | 0 | 1 | 0 | 0 | 7 | 3 | 6 |
| 8106 | 1 | 4 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 11 | ... | 0 | 6 | 7 | 0 | 2 | 0 | 0 | 1 | 3 | 6 |
| 8107 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 10 | ... | 0 | 6 | 7 | 0 | 2 | 0 | 0 | 1 | 5 | 6 |
| 8108 | 0 | 4 | 1 | 4 | 1 | 6 | 0 | 0 | 0 | 8 | ... | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 4 | 3 | 6 |
| 8109 | 1 | 1 | 0 | 2 | 1 | 3 | 0 | 1 | 1 | 4 | ... | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 1 | 1 |
| 8110 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 11 | ... | 0 | 6 | 7 | 0 | 2 | 0 | 0 | 1 | 3 | 6 |
| 8111 | 1 | 4 | 0 | 2 | 1 | 3 | 0 | 1 | 1 | 3 | ... | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 4 | 1 | 1 |
| 8112 | 1 | 4 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 11 | ... | 0 | 6 | 7 | 0 | 1 | 0 | 0 | 8 | 3 | 6 |
| 8113 | 0 | 4 | 1 | 4 | 1 | 6 | 0 | 0 | 0 | 8 | ... | 3 | 2 | 1 | 0 | 0 | 0 | 1 | 4 | 3 | 3 |
| 8114 | 0 | 3 | 1 | 8 | 1 | 8 | 1 | 0 | 1 | 10 | ... | 2 | 7 | 8 | 0 | 0 | 2 | 4 | 4 | 5 | 3 |
| 8115 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 10 | ... | 0 | 6 | 7 | 0 | 2 | 0 | 0 | 6 | 3 | 6 |
| 8116 | 0 | 4 | 1 | 0 | 1 | 7 | 0 | 0 | 0 | 8 | ... | 3 | 2 | 0 | 0 | 0 | 0 | 1 | 4 | 3 | 6 |
| 8117 | 0 | 4 | 0 | 4 | 1 | 6 | 0 | 0 | 0 | 8 | ... | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 4 | 3 | 3 |
| 8118 | 0 | 4 | 1 | 0 | 1 | 4 | 0 | 0 | 0 | 8 | ... | 0 | 2 | 0 | 0 | 0 | 0 | 1 | 4 | 3 | 3 |
| 8119 | 1 | 4 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 10 | ... | 0 | 6 | 7 | 0 | 2 | 0 | 0 | 8 | 5 | 6 |
| 8120 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 10 | ... | 0 | 6 | 7 | 0 | 1 | 0 | 0 | 8 | 3 | 6 |
| 8121 | 1 | 3 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 1 | ... | 0 | 6 | 7 | 0 | 2 | 0 | 0 | 8 | 5 | 6 |
| 8122 | 0 | 4 | 1 | 0 | 1 | 6 | 0 | 0 | 0 | 8 | ... | 3 | 0 | 0 | 0 | 0 | 0 | 1 | 4 | 3 | 6 |
| 8123 | 1 | 0 | 0 | 0 | 1 | 3 | 1 | 0 | 1 | 10 | ... | 0 | 6 | 7 | 0 | 2 | 0 | 0 | 6 | 5 | 6 |
8124 rows × 23 columns
In [40]:
map_dic
Out[40]:
{0: {0: 1, 1: 0},
1: {'b': 1, 'c': 5, 'f': 3, 'k': 4, 's': 2, 'x': 0},
2: {'f': 2, 'g': 3, 's': 0, 'y': 1},
3: {'b': 6,
'c': 8,
'e': 4,
'g': 3,
'n': 0,
'p': 5,
'r': 9,
'u': 7,
'w': 2,
'y': 1},
4: {'f': 1, 't': 0},
5: {'a': 1, 'c': 5, 'f': 4, 'l': 2, 'm': 8, 'n': 3, 'p': 0, 's': 7, 'y': 6},
6: {'a': 1, 'f': 0},
7: {'c': 0, 'w': 1},
8: {'b': 1, 'n': 0},
9: {'b': 8,
'e': 7,
'g': 2,
'h': 5,
'k': 0,
'n': 1,
'o': 11,
'p': 3,
'r': 9,
'u': 6,
'w': 4,
'y': 10},
10: {'e': 0, 't': 1},
11: {'?': 4, 'b': 2, 'c': 1, 'e': 0, 'r': 3},
12: {'f': 1, 'k': 2, 's': 0, 'y': 3},
13: {'f': 1, 'k': 3, 's': 0, 'y': 2},
14: {'b': 4, 'c': 7, 'e': 5, 'g': 1, 'n': 3, 'o': 6, 'p': 2, 'w': 0, 'y': 8},
15: {'b': 3, 'c': 8, 'e': 5, 'g': 2, 'n': 4, 'o': 7, 'p': 1, 'w': 0, 'y': 6},
16: {'p': 0},
17: {'n': 1, 'o': 2, 'w': 0, 'y': 3},
18: {'n': 2, 'o': 0, 't': 1},
19: {'e': 1, 'f': 3, 'l': 2, 'n': 4, 'p': 0},
20: {'b': 8, 'h': 3, 'k': 0, 'n': 1, 'o': 6, 'r': 5, 'u': 2, 'w': 4, 'y': 7},
21: {'a': 2, 'c': 5, 'n': 1, 's': 0, 'v': 3, 'y': 4},
22: {'d': 3, 'g': 1, 'l': 6, 'm': 2, 'p': 4, 'u': 0, 'w': 5}}
In [41]:
df.describe()
Out[41]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | ... | 8124.000000 | 8124.000000 | 8124.000000 | 8124.0 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 |
| mean | 0.517971 | 1.637617 | 0.971935 | 2.093550 | 0.584441 | 3.589365 | 0.025849 | 0.161497 | 0.690793 | 4.274249 | ... | 0.994584 | 1.149680 | 1.061546 | 0.0 | 0.038405 | 0.082718 | 0.696209 | 2.200886 | 2.687839 | 2.794682 |
| std | 0.499708 | 1.588969 | 0.775534 | 1.744794 | 0.492848 | 1.557709 | 0.158695 | 0.368011 | 0.462195 | 2.669652 | ... | 1.333097 | 1.563541 | 1.648595 | 0.0 | 0.257837 | 0.291116 | 0.786930 | 1.742226 | 1.381200 | 1.617549 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 3.000000 | 0.000000 | 0.000000 | 0.000000 | 2.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 1.000000 | 2.000000 | 1.000000 |
| 50% | 1.000000 | 1.000000 | 1.000000 | 2.000000 | 1.000000 | 3.000000 | 0.000000 | 0.000000 | 1.000000 | 4.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 1.000000 | 3.000000 | 3.000000 | 3.000000 |
| 75% | 1.000000 | 3.000000 | 2.000000 | 3.000000 | 1.000000 | 4.000000 | 0.000000 | 0.000000 | 1.000000 | 6.000000 | ... | 3.000000 | 2.000000 | 1.000000 | 0.0 | 0.000000 | 0.000000 | 1.000000 | 4.000000 | 4.000000 | 4.000000 |
| max | 1.000000 | 5.000000 | 3.000000 | 9.000000 | 1.000000 | 8.000000 | 1.000000 | 1.000000 | 1.000000 | 11.000000 | ... | 3.000000 | 8.000000 | 8.000000 | 0.0 | 3.000000 | 2.000000 | 4.000000 | 8.000000 | 5.000000 | 6.000000 |
8 rows × 23 columns
7) 각 컬럼별 Normalization¶
In [42]:
for i in range(1, num_columns):
unique_array = df[i].unique()
N = len(unique_array)
map_dic_sub = {}
for j in range(N):
if j == 0:
map_dic_sub[j] = 0
else:
map_dic_sub[j] = j / float(N - 1)
df[i] = df[i].map(map_dic_sub)
df
Out[42]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0.0 | 0.000000 | 0.000000 | 0.0 | 0.000 | 0.0 | 0.0 | 0.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.0 | 0.000000 |
| 1 | 1 | 0.0 | 0.000000 | 0.111111 | 0.0 | 0.125 | 0.0 | 0.0 | 1.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.2 | 0.166667 |
| 2 | 1 | 0.2 | 0.000000 | 0.222222 | 0.0 | 0.250 | 0.0 | 0.0 | 1.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.2 | 0.333333 |
| 3 | 0 | 0.0 | 0.333333 | 0.222222 | 0.0 | 0.000 | 0.0 | 0.0 | 0.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.0 | 0.000000 |
| 4 | 1 | 0.0 | 0.000000 | 0.333333 | 1.0 | 0.375 | 0.0 | 1.0 | 1.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.25 | 0.125 | 0.4 | 0.166667 |
| 5 | 1 | 0.0 | 0.333333 | 0.111111 | 0.0 | 0.125 | 0.0 | 0.0 | 1.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.2 | 0.166667 |
| 6 | 1 | 0.2 | 0.000000 | 0.222222 | 0.0 | 0.125 | 0.0 | 0.0 | 1.0 | 0.181818 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.2 | 0.333333 |
| 7 | 1 | 0.2 | 0.333333 | 0.222222 | 0.0 | 0.250 | 0.0 | 0.0 | 1.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.0 | 0.333333 |
| 8 | 0 | 0.0 | 0.333333 | 0.222222 | 0.0 | 0.000 | 0.0 | 0.0 | 0.0 | 0.272727 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.6 | 0.166667 |
| 9 | 1 | 0.2 | 0.000000 | 0.111111 | 0.0 | 0.125 | 0.0 | 0.0 | 1.0 | 0.181818 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.0 | 0.333333 |
| 10 | 1 | 0.0 | 0.333333 | 0.111111 | 0.0 | 0.250 | 0.0 | 0.0 | 1.0 | 0.181818 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.2 | 0.166667 |
| 11 | 1 | 0.0 | 0.333333 | 0.111111 | 0.0 | 0.125 | 0.0 | 0.0 | 1.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.0 | 0.333333 |
| 12 | 1 | 0.2 | 0.000000 | 0.111111 | 0.0 | 0.125 | 0.0 | 0.0 | 1.0 | 0.363636 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.0 | 0.166667 |
| 13 | 0 | 0.0 | 0.333333 | 0.222222 | 0.0 | 0.000 | 0.0 | 0.0 | 0.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.6 | 0.000000 |
| 14 | 1 | 0.0 | 0.666667 | 0.000000 | 1.0 | 0.375 | 0.0 | 1.0 | 1.0 | 0.090909 | ... | 0.333333 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.25 | 0.000 | 0.4 | 0.166667 |
| 15 | 1 | 0.4 | 0.666667 | 0.333333 | 1.0 | 0.375 | 0.0 | 0.0 | 0.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.8 | 0.000000 |
| 16 | 1 | 0.6 | 0.666667 | 0.222222 | 1.0 | 0.375 | 0.0 | 1.0 | 1.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.25 | 0.125 | 0.4 | 0.166667 |
| 17 | 0 | 0.0 | 0.000000 | 0.000000 | 0.0 | 0.000 | 0.0 | 0.0 | 0.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.0 | 0.166667 |
| 18 | 0 | 0.0 | 0.333333 | 0.222222 | 0.0 | 0.000 | 0.0 | 0.0 | 0.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.0 | 0.000000 |
| 19 | 0 | 0.0 | 0.000000 | 0.000000 | 0.0 | 0.000 | 0.0 | 0.0 | 0.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.0 | 0.000000 |
| 20 | 1 | 0.2 | 0.000000 | 0.111111 | 0.0 | 0.125 | 0.0 | 0.0 | 1.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.0 | 0.333333 |
| 21 | 0 | 0.0 | 0.333333 | 0.000000 | 0.0 | 0.000 | 0.0 | 0.0 | 0.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.6 | 0.166667 |
| 22 | 1 | 0.2 | 0.333333 | 0.111111 | 0.0 | 0.250 | 0.0 | 0.0 | 1.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.0 | 0.333333 |
| 23 | 1 | 0.2 | 0.333333 | 0.222222 | 0.0 | 0.125 | 0.0 | 0.0 | 1.0 | 0.363636 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.2 | 0.333333 |
| 24 | 1 | 0.2 | 0.000000 | 0.222222 | 0.0 | 0.250 | 0.0 | 0.0 | 1.0 | 0.181818 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.0 | 0.333333 |
| 25 | 0 | 0.6 | 0.000000 | 0.222222 | 0.0 | 0.000 | 0.0 | 0.0 | 0.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.6 | 0.166667 |
| 26 | 1 | 0.0 | 0.333333 | 0.111111 | 0.0 | 0.125 | 0.0 | 0.0 | 1.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.2 | 0.333333 |
| 27 | 1 | 0.0 | 0.333333 | 0.222222 | 0.0 | 0.250 | 0.0 | 0.0 | 1.0 | 0.363636 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.2 | 0.333333 |
| 28 | 1 | 0.6 | 0.666667 | 0.000000 | 1.0 | 0.375 | 0.0 | 0.0 | 0.0 | 0.000000 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.000 | 0.8 | 0.000000 |
| 29 | 1 | 0.0 | 0.000000 | 0.111111 | 0.0 | 0.125 | 0.0 | 1.0 | 0.0 | 0.090909 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.00 | 0.125 | 0.6 | 0.500000 |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 8094 | 1 | 0.2 | 0.000000 | 0.333333 | 1.0 | 0.375 | 0.0 | 1.0 | 1.0 | 0.181818 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.5 | 0.00 | 0.500 | 0.2 | 0.166667 |
| 8095 | 0 | 0.0 | 0.333333 | 0.888889 | 1.0 | 1.000 | 0.0 | 0.0 | 1.0 | 0.909091 | ... | 0.666667 | 0.875 | 1.000 | 0 | 0.000000 | 1.0 | 1.00 | 0.500 | 1.0 | 0.500000 |
| 8096 | 1 | 0.8 | 0.666667 | 0.222222 | 1.0 | 0.375 | 0.0 | 1.0 | 1.0 | 0.363636 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.5 | 0.00 | 0.500 | 0.2 | 0.166667 |
| 8097 | 0 | 0.8 | 0.333333 | 0.000000 | 1.0 | 0.875 | 0.0 | 0.0 | 0.0 | 0.727273 | ... | 1.000000 | 0.250 | 0.125 | 0 | 0.000000 | 0.0 | 0.25 | 0.500 | 0.6 | 1.000000 |
| 8098 | 0 | 0.8 | 0.000000 | 0.444444 | 1.0 | 0.750 | 0.0 | 0.0 | 0.0 | 0.727273 | ... | 1.000000 | 0.000 | 0.125 | 0 | 0.000000 | 0.0 | 0.25 | 0.500 | 0.6 | 0.500000 |
| 8099 | 1 | 0.8 | 0.666667 | 0.222222 | 1.0 | 0.375 | 0.0 | 1.0 | 1.0 | 0.363636 | ... | 1.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.5 | 0.00 | 0.500 | 0.0 | 0.166667 |
| 8100 | 1 | 0.6 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 1.000000 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.333333 | 0.0 | 0.00 | 1.000 | 0.6 | 1.000000 |
| 8101 | 0 | 0.8 | 0.000000 | 0.444444 | 1.0 | 0.875 | 0.0 | 0.0 | 0.0 | 0.727273 | ... | 0.000000 | 0.250 | 0.000 | 0 | 0.000000 | 0.0 | 0.25 | 0.500 | 0.6 | 0.666667 |
| 8102 | 1 | 0.0 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.333333 | 0.0 | 0.00 | 0.125 | 1.0 | 1.000000 |
| 8103 | 1 | 0.8 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.333333 | 0.0 | 0.00 | 0.750 | 1.0 | 1.000000 |
| 8104 | 1 | 0.8 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.666667 | 0.0 | 0.00 | 0.125 | 0.6 | 1.000000 |
| 8105 | 1 | 0.8 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.333333 | 0.0 | 0.00 | 0.875 | 0.6 | 1.000000 |
| 8106 | 1 | 0.8 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 1.000000 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.666667 | 0.0 | 0.00 | 0.125 | 0.6 | 1.000000 |
| 8107 | 1 | 0.0 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.666667 | 0.0 | 0.00 | 0.125 | 1.0 | 1.000000 |
| 8108 | 0 | 0.8 | 0.333333 | 0.444444 | 1.0 | 0.750 | 0.0 | 0.0 | 0.0 | 0.727273 | ... | 0.000000 | 0.250 | 0.000 | 0 | 0.000000 | 0.0 | 0.25 | 0.500 | 0.6 | 1.000000 |
| 8109 | 1 | 0.2 | 0.000000 | 0.222222 | 1.0 | 0.375 | 0.0 | 1.0 | 1.0 | 0.363636 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.5 | 0.00 | 0.500 | 0.2 | 0.166667 |
| 8110 | 1 | 0.0 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 1.000000 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.666667 | 0.0 | 0.00 | 0.125 | 0.6 | 1.000000 |
| 8111 | 1 | 0.8 | 0.000000 | 0.222222 | 1.0 | 0.375 | 0.0 | 1.0 | 1.0 | 0.272727 | ... | 0.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.5 | 0.00 | 0.500 | 0.2 | 0.166667 |
| 8112 | 1 | 0.8 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 1.000000 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.333333 | 0.0 | 0.00 | 1.000 | 0.6 | 1.000000 |
| 8113 | 0 | 0.8 | 0.333333 | 0.444444 | 1.0 | 0.750 | 0.0 | 0.0 | 0.0 | 0.727273 | ... | 1.000000 | 0.250 | 0.125 | 0 | 0.000000 | 0.0 | 0.25 | 0.500 | 0.6 | 0.500000 |
| 8114 | 0 | 0.6 | 0.333333 | 0.888889 | 1.0 | 1.000 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.666667 | 0.875 | 1.000 | 0 | 0.000000 | 1.0 | 1.00 | 0.500 | 1.0 | 0.500000 |
| 8115 | 1 | 0.0 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.666667 | 0.0 | 0.00 | 0.750 | 0.6 | 1.000000 |
| 8116 | 0 | 0.8 | 0.333333 | 0.000000 | 1.0 | 0.875 | 0.0 | 0.0 | 0.0 | 0.727273 | ... | 1.000000 | 0.250 | 0.000 | 0 | 0.000000 | 0.0 | 0.25 | 0.500 | 0.6 | 1.000000 |
| 8117 | 0 | 0.8 | 0.000000 | 0.444444 | 1.0 | 0.750 | 0.0 | 0.0 | 0.0 | 0.727273 | ... | 0.000000 | 0.250 | 0.000 | 0 | 0.000000 | 0.0 | 0.25 | 0.500 | 0.6 | 0.500000 |
| 8118 | 0 | 0.8 | 0.333333 | 0.000000 | 1.0 | 0.500 | 0.0 | 0.0 | 0.0 | 0.727273 | ... | 0.000000 | 0.250 | 0.000 | 0 | 0.000000 | 0.0 | 0.25 | 0.500 | 0.6 | 0.500000 |
| 8119 | 1 | 0.8 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.666667 | 0.0 | 0.00 | 1.000 | 1.0 | 1.000000 |
| 8120 | 1 | 0.0 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.333333 | 0.0 | 0.00 | 1.000 | 0.6 | 1.000000 |
| 8121 | 1 | 0.6 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.090909 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.666667 | 0.0 | 0.00 | 1.000 | 1.0 | 1.000000 |
| 8122 | 0 | 0.8 | 0.333333 | 0.000000 | 1.0 | 0.750 | 0.0 | 0.0 | 0.0 | 0.727273 | ... | 1.000000 | 0.000 | 0.000 | 0 | 0.000000 | 0.0 | 0.25 | 0.500 | 0.6 | 1.000000 |
| 8123 | 1 | 0.0 | 0.000000 | 0.000000 | 1.0 | 0.375 | 1.0 | 0.0 | 1.0 | 0.909091 | ... | 0.000000 | 0.750 | 0.875 | 0 | 0.666667 | 0.0 | 0.00 | 0.750 | 1.0 | 1.000000 |
8124 rows × 23 columns
In [49]:
df.describe()
Out[49]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | ... | 8124.000000 | 8124.000000 | 8124.000000 | 8124.0 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 | 8124.000000 |
| mean | 0.517971 | 0.327523 | 0.323978 | 0.232617 | 0.584441 | 0.448671 | 0.025849 | 0.161497 | 0.690793 | 0.388568 | ... | 0.331528 | 0.143710 | 0.132693 | 0.0 | 0.012802 | 0.041359 | 0.174052 | 0.275111 | 0.537568 | 0.465780 |
| std | 0.499708 | 0.317794 | 0.258511 | 0.193866 | 0.492848 | 0.194714 | 0.158695 | 0.368011 | 0.462195 | 0.242696 | ... | 0.444366 | 0.195443 | 0.206074 | 0.0 | 0.085946 | 0.145558 | 0.196733 | 0.217778 | 0.276240 | 0.269591 |
| min | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.375000 | 0.000000 | 0.000000 | 0.000000 | 0.181818 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.125000 | 0.400000 | 0.166667 |
| 50% | 1.000000 | 0.200000 | 0.333333 | 0.222222 | 1.000000 | 0.375000 | 0.000000 | 0.000000 | 1.000000 | 0.363636 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.250000 | 0.375000 | 0.600000 | 0.500000 |
| 75% | 1.000000 | 0.600000 | 0.666667 | 0.333333 | 1.000000 | 0.500000 | 0.000000 | 0.000000 | 1.000000 | 0.545455 | ... | 1.000000 | 0.250000 | 0.125000 | 0.0 | 0.000000 | 0.000000 | 0.250000 | 0.500000 | 0.800000 | 0.666667 |
| max | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | ... | 1.000000 | 1.000000 | 1.000000 | 0.0 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 |
8 rows × 23 columns
8) Edible Mushrooms과 Poisonous Mushrooms 의 두 개의 그룹핑 작업 및 각 그룹별 Boxplot 그리기¶
- Typical Scematic Box Plot
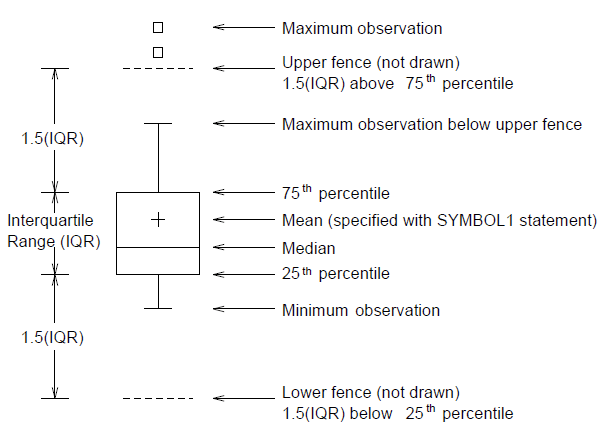
In [50]:
df_edible = df[df[0] == 0] # 0: edible
df_poisonous = df[df[0] == 1] # 1: poisonous
In [51]:
df_edible.describe()
Out[51]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 3916.0 | 3916.000000 | 3916.000000 | 3916.000000 | 3916.000000 | 3916.000000 | 3916.000000 | 3916.000000 | 3916.000000 | 3916.000000 | ... | 3916.000000 | 3916.000000 | 3916.000000 | 3916.0 | 3916.000000 | 3916.000000 | 3916.000000 | 3916.000000 | 3916.000000 | 3916.000000 |
| mean | 0.0 | 0.364454 | 0.278515 | 0.241062 | 0.840654 | 0.566139 | 0.004597 | 0.028601 | 0.432074 | 0.492432 | ... | 0.576779 | 0.189351 | 0.153728 | 0.0 | 0.002043 | 0.018386 | 0.287538 | 0.401685 | 0.582022 | 0.519237 |
| std | 0.0 | 0.331386 | 0.243088 | 0.195300 | 0.366045 | 0.213075 | 0.067650 | 0.166702 | 0.495428 | 0.236910 | ... | 0.481331 | 0.192389 | 0.195711 | 0.0 | 0.045158 | 0.115996 | 0.193596 | 0.138892 | 0.205576 | 0.288464 |
| min | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 0.0 | 0.000000 | 0.000000 | 0.000000 | 1.000000 | 0.500000 | 0.000000 | 0.000000 | 0.000000 | 0.272727 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.250000 | 0.375000 | 0.600000 | 0.166667 |
| 50% | 0.0 | 0.600000 | 0.333333 | 0.222222 | 1.000000 | 0.500000 | 0.000000 | 0.000000 | 0.000000 | 0.454545 | ... | 1.000000 | 0.250000 | 0.125000 | 0.0 | 0.000000 | 0.000000 | 0.250000 | 0.375000 | 0.600000 | 0.500000 |
| 75% | 0.0 | 0.600000 | 0.333333 | 0.444444 | 1.000000 | 0.750000 | 0.000000 | 0.000000 | 1.000000 | 0.727273 | ... | 1.000000 | 0.250000 | 0.125000 | 0.0 | 0.000000 | 0.000000 | 0.500000 | 0.500000 | 0.600000 | 0.666667 |
| max | 0.0 | 1.000000 | 1.000000 | 0.888889 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | 0.909091 | ... | 1.000000 | 1.000000 | 1.000000 | 0.0 | 1.000000 | 1.000000 | 1.000000 | 0.625000 | 1.000000 | 1.000000 |
8 rows × 23 columns
In [52]:
import matplotlib.pyplot as plt
fig, ax = plt.subplots()
fig.set_size_inches(15, 4)
df_edible.boxplot(ax=ax)
plt.show()
/Users/yhhan/anaconda/lib/python2.7/site-packages/ipykernel/__main__.py:4: FutureWarning: The default value for 'return_type' will change to 'axes' in a future release. To use the future behavior now, set return_type='axes'. To keep the previous behavior and silence this warning, set return_type='dict'.
In [53]:
df_poisonous.describe()
Out[53]:
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | 21 | 22 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 4208.0 | 4208.000000 | 4208.000000 | 4208.000000 | 4208.000000 | 4208.000000 | 4208.000000 | 4208.000000 | 4208.000000 | 4208.000000 | ... | 4208.000000 | 4208.000000 | 4208.000000 | 4208.0 | 4208.000000 | 4208.000000 | 4208.000000 | 4208.000000 | 4208.000000 | 4208.000000 |
| mean | 1.0 | 0.293156 | 0.366286 | 0.224757 | 0.346008 | 0.339354 | 0.045627 | 0.285171 | 0.931559 | 0.291912 | ... | 0.103295 | 0.101236 | 0.113118 | 0.0 | 0.022814 | 0.062738 | 0.068441 | 0.157319 | 0.496198 | 0.416033 |
| std | 0.0 | 0.300591 | 0.265197 | 0.192212 | 0.475752 | 0.078467 | 0.208700 | 0.451550 | 0.252531 | 0.205210 | ... | 0.239945 | 0.188588 | 0.213440 | 0.0 | 0.110257 | 0.165648 | 0.129258 | 0.211734 | 0.323201 | 0.240334 |
| min | 1.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.125000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
| 25% | 1.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.375000 | 0.000000 | 0.000000 | 1.000000 | 0.090909 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.200000 | 0.166667 |
| 50% | 1.0 | 0.200000 | 0.333333 | 0.222222 | 0.000000 | 0.375000 | 0.000000 | 0.000000 | 1.000000 | 0.272727 | ... | 0.000000 | 0.000000 | 0.000000 | 0.0 | 0.000000 | 0.000000 | 0.000000 | 0.125000 | 0.600000 | 0.500000 |
| 75% | 1.0 | 0.600000 | 0.666667 | 0.333333 | 1.000000 | 0.375000 | 0.000000 | 1.000000 | 1.000000 | 0.363636 | ... | 0.000000 | 0.125000 | 0.125000 | 0.0 | 0.000000 | 0.000000 | 0.250000 | 0.125000 | 0.800000 | 0.500000 |
| max | 1.0 | 0.800000 | 0.666667 | 1.000000 | 1.000000 | 0.375000 | 1.000000 | 1.000000 | 1.000000 | 1.000000 | ... | 1.000000 | 0.750000 | 0.875000 | 0.0 | 0.666667 | 0.500000 | 0.750000 | 1.000000 | 1.000000 | 1.000000 |
8 rows × 23 columns
In [54]:
fig, ax = plt.subplots()
fig.set_size_inches(15, 4)
df_poisonous.boxplot(ax=ax)
plt.show()
/Users/yhhan/anaconda/lib/python2.7/site-packages/ipykernel/__main__.py:3: FutureWarning: The default value for 'return_type' will change to 'axes' in a future release. To use the future behavior now, set return_type='axes'. To keep the previous behavior and silence this warning, set return_type='dict'. app.launch_new_instance()
9) 두 그룹간에 확연하게 차이를 보이는 속성들에 대한 EDA 작업¶
- 속성 4 (bruises: bruises=t (0), no=f (1))
In [55]:
map_dic[4]
Out[55]:
{'f': 1, 't': 0}
In [56]:
df_edible_4 = df_edible[[0, 4]]
df_edible_4.describe()
Out[56]:
| 0 | 4 | |
|---|---|---|
| count | 3916.0 | 3916.000000 |
| mean | 0.0 | 0.840654 |
| std | 0.0 | 0.366045 |
| min | 0.0 | 0.000000 |
| 25% | 0.0 | 1.000000 |
| 50% | 0.0 | 1.000000 |
| 75% | 0.0 | 1.000000 |
| max | 0.0 | 1.000000 |
In [57]:
df_edible_groupby_4 = df_edible_4.groupby(4)
df_edible_groupby_4.count()
Out[57]:
| 0 | |
|---|---|
| 4 | |
| 0.0 | 624 |
| 1.0 | 3292 |
In [58]:
df_poisonous_4 = df_poisonous[[0, 4]]
df_poisonous_4.describe()
Out[58]:
| 0 | 4 | |
|---|---|---|
| count | 4208.0 | 4208.000000 |
| mean | 1.0 | 0.346008 |
| std | 0.0 | 0.475752 |
| min | 1.0 | 0.000000 |
| 25% | 1.0 | 0.000000 |
| 50% | 1.0 | 0.000000 |
| 75% | 1.0 | 1.000000 |
| max | 1.0 | 1.000000 |
In [59]:
df_poisonous_groupby_4 = df_poisonous_4.groupby(4)
df_poisonous_groupby_4.count()
Out[59]:
| 0 | |
|---|---|
| 4 | |
| 0.0 | 2752 |
| 1.0 | 1456 |
- 속성 5 (odor: almond=a, anise=l, creosote=c, fishy=y, foul=f, musty=m, none=n, pungent=p, spicy=s)
In [60]:
map_dic[5]
Out[60]:
{'a': 1, 'c': 5, 'f': 4, 'l': 2, 'm': 8, 'n': 3, 'p': 0, 's': 7, 'y': 6}
In [61]:
df_edible_5 = df_edible[[0, 5]]
df_edible_5.describe()
Out[61]:
| 0 | 5 | |
|---|---|---|
| count | 3916.0 | 3916.000000 |
| mean | 0.0 | 0.566139 |
| std | 0.0 | 0.213075 |
| min | 0.0 | 0.000000 |
| 25% | 0.0 | 0.500000 |
| 50% | 0.0 | 0.500000 |
| 75% | 0.0 | 0.750000 |
| max | 0.0 | 1.000000 |
In [62]:
df_edible_groupby_5 = df_edible_5.groupby(5)
df_edible_groupby_5.count()
Out[62]:
| 0 | |
|---|---|
| 5 | |
| 0.000 | 256 |
| 0.375 | 120 |
| 0.500 | 2160 |
| 0.625 | 192 |
| 0.750 | 576 |
| 0.875 | 576 |
| 1.000 | 36 |
In [63]:
df_poisonous_5 = df_poisonous[[0, 5]]
df_poisonous_5.describe()
Out[63]:
| 0 | 5 | |
|---|---|---|
| count | 4208.0 | 4208.000000 |
| mean | 1.0 | 0.339354 |
| std | 0.0 | 0.078467 |
| min | 1.0 | 0.125000 |
| 25% | 1.0 | 0.375000 |
| 50% | 1.0 | 0.375000 |
| 75% | 1.0 | 0.375000 |
| max | 1.0 | 0.375000 |
In [64]:
df_poisonous_groupby_5 = df_poisonous_5.groupby(5)
df_poisonous_groupby_5.count()
Out[64]:
| 0 | |
|---|---|
| 5 | |
| 0.125 | 400 |
| 0.250 | 400 |
| 0.375 | 3408 |
- 속성 16 (veil-type: partial=p, universal=u)
In [65]:
map_dic[16]
Out[65]:
{'p': 0}
- 속성 17 (veil-color: brown=n, orange=o, white=w, yellow=y)
In [66]:
map_dic[17]
Out[66]:
{'n': 1, 'o': 2, 'w': 0, 'y': 3}
In [67]:
df_edible_17 = df_edible[[0, 17]]
df_edible_17.describe()
Out[67]:
| 0 | 17 | |
|---|---|---|
| count | 3916.0 | 3916.000000 |
| mean | 0.0 | 0.002043 |
| std | 0.0 | 0.045158 |
| min | 0.0 | 0.000000 |
| 25% | 0.0 | 0.000000 |
| 50% | 0.0 | 0.000000 |
| 75% | 0.0 | 0.000000 |
| max | 0.0 | 1.000000 |
In [68]:
df_edible_groupby_17 = df_edible_17.groupby(17)
df_edible_groupby_17.count()
Out[68]:
| 0 | |
|---|---|
| 17 | |
| 0.0 | 3908 |
| 1.0 | 8 |
In [69]:
df_poisonous_17 = df_poisonous[[0, 17]]
df_poisonous_17.describe()
Out[69]:
| 0 | 17 | |
|---|---|---|
| count | 4208.0 | 4208.000000 |
| mean | 1.0 | 0.022814 |
| std | 0.0 | 0.110257 |
| min | 1.0 | 0.000000 |
| 25% | 1.0 | 0.000000 |
| 50% | 1.0 | 0.000000 |
| 75% | 1.0 | 0.000000 |
| max | 1.0 | 0.666667 |
In [70]:
df_poisonous_groupby_17 = df_poisonous_17.groupby(17)
df_poisonous_groupby_17.count()
Out[70]:
| 0 | |
|---|---|
| 17 | |
| 0.000000 | 4016 |
| 0.333333 | 96 |
| 0.666667 | 96 |