Random Walks in the Matrix¶
Imagine standing at the center of a sphere, with 12 equally-sized spheres all around you, at the corners of a cuboctahedron. Each time you step into a random neighboring sphere, you're in the same circumstance, of having a choice of twelve nextdoor neighbors.
Have you seen random walks on chess or checker boards? Just take the checker pattern and extend it indefinitely around an arbitrary square marked with a lamp post.
Our little man has maybe had too much to drink and is moving randomly from one square to the next, but only across an edge, not through a diagonal. On a square grid, he has four neighbors to select from. On an hexagonal grid, he would have six.
Back to our space-filling grid, our cells are rhombic dodecahedron shaped (twelve diamond faces). We're free to hop at random, leaving a trace over time, of what our pathway has been. This concept of a pathway applies regardless of which grid we're imagining.
What we're going to find is that four random walkers, setting out from the same origin (lamp post) will define the four CCP sphere corners of a random tetrahedron. That tetrahedron will have a whole number volume relative to the unit tetrahedron of four CCP balls and volume one.
Quadrays¶
To make our pathway through the CCP (the name of this sphere packing pattern) easy to compute and keep track of, we're going to want to label all the spheres with vectorial coordinates.
We will find it convenient to do so using Quadrays, an interface bolted on XYZ and easy to reason about independently of XYZ. In this game of vectors, the origin sphere will be labeled (0,0,0,0) and the twelve around it will be the unique permutations of (2,1,1,0).
Each time our random walker takes a step, he has one of twelve vectors to choose from. Adding that vector to his current position takes him to a next position and so on, onward along a pathway.
Linear combinations of the four basis rays (lets use that shoptalk) map to any point in space, as expressed by 4-tuples.
At least one of the four rays, the one not bordering a specified point's quadrant, will not contribute to wayfinding. In canonical form, at least one element of the 4-tuple will be zero, the others non-negative.
Qvectors are additive and scalable, have length and direction, according to the usual rules of vector algebra.
Two way conversion from to XYZ is assurred, given Qvectors come with the one canonical representation per point, just as XYZ does.
Quadrays may be seen as "syntactic sugar" atop XYZ, an API. Or vice versa.
from qrays import Qvector
from math import sqrt
Is it bizarre to the point of macabre to have our basis rays not also be of unit length?
The tetrahedron cage is more important: it's edges should be 1 exactly (or 2 if measured in sphere radii).
Our context is the IVM (= CCP = FCC), a closest packed spheres pattern well known to mathematicians, but by other names.
a = Qvector((1,0,0,0))
a.length()
0.6123724356957945
b = Qvector((0,1,0,0))
b.length()
0.6123724356957945
To subtract is to add the additive inverse of, which is the Qvector rotated to point oppositely. (1,0,0,0), negated, is (0,1,1,1).
The result of a subtraction is another Qvector, and its length will be the length separating the "arrow tips" of the two subtracted.
(a-b).length() # the edge of our base tetrahedron
1.0
IVM Vectors¶
Two of any quadray, plus one each of two others, plus zero, will always point to one of the twelve directions surrounding any sphere in the IVM (CCP).
In other words, every unique permutation of {2, 1, 1, 0} will be a vector from the origin to one of the corners of a cuboctahedron, a ball center one sphere diameter distant.
Our random walks will randomly choose one of these twelve directions at each turn to play. Lets call these "moves" (as in chess moves).
from itertools import permutations
from random import choice
moves = [Qvector(p) for p in set(permutations((2,1,1,0)))]
moves
[ivm_vector(a=0, b=1, c=1, d=2), ivm_vector(a=1, b=0, c=1, d=2), ivm_vector(a=2, b=0, c=1, d=1), ivm_vector(a=0, b=2, c=1, d=1), ivm_vector(a=0, b=1, c=2, d=1), ivm_vector(a=1, b=2, c=1, d=0), ivm_vector(a=1, b=1, c=2, d=0), ivm_vector(a=2, b=1, c=1, d=0), ivm_vector(a=1, b=0, c=2, d=1), ivm_vector(a=1, b=2, c=0, d=1), ivm_vector(a=2, b=1, c=0, d=1), ivm_vector(a=1, b=1, c=0, d=2)]
def random_walk(start, steps):
end = start
for _ in range(steps):
end += choice(moves)
return end
vA = random_walk(Qvector((0,0,0,0)), 1000)
vB = random_walk(Qvector((0,0,0,0)), 1000)
vC = random_walk(Qvector((0,0,0,0)), 1000)
vD = random_walk(Qvector((0,0,0,0)), 1000)
import tetravolume as tv
c = Qvector((0,0,1,0))
d = Qvector((0,0,0,1))
ab = (a-b).length()
ac = (a-c).length()
ad = (a-d).length()
bc = (b-c).length()
cd = (c-d).length()
bd = (b-d).length()
t = tv.Tetrahedron(ab,ac,ad,bc,cd,ad)
t.ivm_volume()
1.0
t.xyz_volume()
0.9428090415820635
(a.xyz,
b.xyz,
c.xyz,
d.xyz)
(xyz_vector(x=mpfr('0.35355339059327373'), y=mpfr('0.35355339059327373'), z=mpfr('0.35355339059327373')),
xyz_vector(x=mpfr('-0.35355339059327373'), y=mpfr('-0.35355339059327373'), z=mpfr('0.35355339059327373')),
xyz_vector(x=mpfr('-0.35355339059327373'), y=mpfr('0.35355339059327373'), z=mpfr('-0.35355339059327373')),
xyz_vector(x=mpfr('0.35355339059327373'), y=mpfr('-0.35355339059327373'), z=mpfr('-0.35355339059327373')))
Now lets get the volume using our random IVM corners.
ab = (vA-vB).length()
ac = (vA-vC).length()
ad = (vA-vD).length()
bc = (vB-vC).length()
cd = (vC-vD).length()
bd = (vB-vD).length()
t = tv.Tetrahedron(ab,ac,ad,bc,cd,bd)
t.ivm_volume()
6958.999999999965
Just keep re-running the cell below to appreciate how we only get whole number tetravolumes, for all of these random tetrahedrons. Within floating point rounding error.
vA = random_walk(Qvector((0,0,0,0)), 1000)
vB = random_walk(Qvector((0,0,0,0)), 1000)
vC = random_walk(Qvector((0,0,0,0)), 1000)
vD = random_walk(Qvector((0,0,0,0)), 1000)
ab = (vA-vB).length()
ac = (vA-vC).length()
ad = (vA-vD).length()
bc = (vB-vC).length()
cd = (vC-vD).length()
bd = (vB-vD).length()
t = tv.Tetrahedron(ab,ac,ad,bc,cd,bd)
round(t.ivm_volume(), 8)
3424.0
import gmpy2
from gmpy2 import mpfr
gmpy2.get_context().precision=200
z0 = mpfr('0')
vA = random_walk(Qvector((z0,z0,z0,z0)), 1000)
vB = random_walk(Qvector((z0,z0,z0,z0)), 1000)
vC = random_walk(Qvector((z0,z0,z0,z0)), 1000)
vD = random_walk(Qvector((z0,z0,z0,z0)), 1000)
ab = (vA-vB).length()
ac = (vA-vC).length()
ad = (vA-vD).length()
bc = (vB-vC).length()
cd = (vC-vD).length()
bd = (vB-vD).length()
t = tv.Tetrahedron(ab,ac,ad,bc,cd,bd)
t.ivm_volume()
mpfr('16772.0',200)
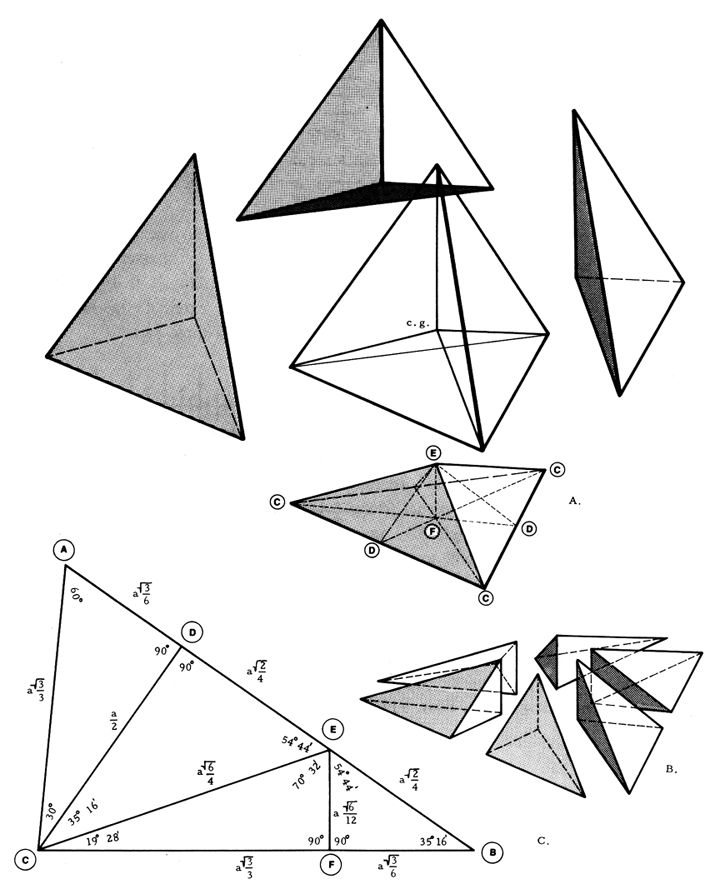
Volume of the A Module¶
The cells below confirm that our base tetrahedron of unit volume, fractionates into 24 subvolumes with angles and edges at our finger tips, thanks to the qrays module
amod_E = Qvector((0,0,0,0))
amod_C = b
amod_D = (b + c)/2
amod_F = (b + c + d)/3
# apex E to base F, C, D
amod_EF = amod_F
amod_CE = b
amod_DE = amod_D
# around the base, C, D, E
amod_CF = amod_C - amod_F
amod_CD = amod_C - amod_D
amod_DF = amod_D - amod_F
amod_EF.length() # apex to face center
0.2041241452319315
amod_CE.length() # apex to corner
0.6123724356957945
amod_DE.length() # apex to mid-edge
0.3535533905932738
amod_CF.length() # face center to corner
0.5773502691896258
amod_CD.length() # corner to mid-edge
0.5
amod_DF.length() # mid-edge to face center
0.2886751345948129
lengths = map(lambda v: v.length(),
(amod_EF, amod_CE, amod_DE, amod_CF, amod_CD, amod_DF))
t = tv.Tetrahedron(*lengths)
t.ivm_volume()
0.04166666666666655
And now, in higher precision...¶
one = mpfr('1')
a = Qvector((one, z0, z0, z0))
b = Qvector((z0, one, z0, z0))
c = Qvector((z0, z0, one, z0))
d = Qvector((z0, z0, z0, one))
(a-b).length()
mpfr('1.0',200)
amod_E = Qvector((z0,z0,z0,z0))
amod_C = b
amod_D = (b + c)/mpfr('2')
amod_F = (b + c + d)/mpfr('3')
# apex E to base F, C, D
amod_EF = amod_F
amod_CE = b
amod_DE = amod_D
# around the base, C, D, E
amod_CF = amod_C - amod_F
amod_CD = amod_C - amod_D
amod_DF = amod_D - amod_F
lengths = list(map(lambda v: v.length(),
(amod_EF, amod_CE, amod_DE, amod_CF, amod_CD, amod_DF)))
lengths[0] # checking length
mpfr('0.20412414523193150818310700622549094933049562338805584403605771',200)
gmpy2.sqrt(mpfr('6'))/12 # length of EF in Fig. 913.01 of Synergetics
mpfr('0.20412414523193150818310700622549094933049562338805584403605771',200)
t = tv.Tetrahedron(*lengths)
t.ivm_volume()
mpfr('0.041666666666666666666666666666666666666666666666666666666666835',200)