Course: Building an Effective ML Workflow with scikit-learn¶
Starter code (copy from here: http://bit.ly/first-ml-lesson)¶
import pandas as pd
from sklearn.preprocessing import OneHotEncoder
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.compose import make_column_transformer
from sklearn.pipeline import make_pipeline
cols = ['Parch', 'Fare', 'Embarked', 'Sex', 'Name']
df = pd.read_csv('http://bit.ly/kaggletrain', nrows=10)
X = df[cols]
y = df['Survived']
df_new = pd.read_csv('http://bit.ly/kaggletest', nrows=10)
X_new = df_new[cols]
ohe = OneHotEncoder()
vect = CountVectorizer()
ct = make_column_transformer(
(ohe, ['Embarked', 'Sex']),
(vect, 'Name'),
remainder='passthrough')
logreg = LogisticRegression(solver='liblinear', random_state=1)
pipe = make_pipeline(ct, logreg)
pipe.fit(X, y)
pipe.predict(X_new)
array([0, 1, 0, 0, 1, 0, 1, 0, 1, 0])
Part 5: Handling missing values¶
We want to use "Age" as a feature, but note that it has a missing value (encoded as "NaN"):
df
| PassengerId | Survived | Pclass | Name | Sex | Age | SibSp | Parch | Ticket | Fare | Cabin | Embarked | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 0 | 3 | Braund, Mr. Owen Harris | male | 22.0 | 1 | 0 | A/5 21171 | 7.2500 | NaN | S |
| 1 | 2 | 1 | 1 | Cumings, Mrs. John Bradley (Florence Briggs Th... | female | 38.0 | 1 | 0 | PC 17599 | 71.2833 | C85 | C |
| 2 | 3 | 1 | 3 | Heikkinen, Miss. Laina | female | 26.0 | 0 | 0 | STON/O2. 3101282 | 7.9250 | NaN | S |
| 3 | 4 | 1 | 1 | Futrelle, Mrs. Jacques Heath (Lily May Peel) | female | 35.0 | 1 | 0 | 113803 | 53.1000 | C123 | S |
| 4 | 5 | 0 | 3 | Allen, Mr. William Henry | male | 35.0 | 0 | 0 | 373450 | 8.0500 | NaN | S |
| 5 | 6 | 0 | 3 | Moran, Mr. James | male | NaN | 0 | 0 | 330877 | 8.4583 | NaN | Q |
| 6 | 7 | 0 | 1 | McCarthy, Mr. Timothy J | male | 54.0 | 0 | 0 | 17463 | 51.8625 | E46 | S |
| 7 | 8 | 0 | 3 | Palsson, Master. Gosta Leonard | male | 2.0 | 3 | 1 | 349909 | 21.0750 | NaN | S |
| 8 | 9 | 1 | 3 | Johnson, Mrs. Oscar W (Elisabeth Vilhelmina Berg) | female | 27.0 | 0 | 2 | 347742 | 11.1333 | NaN | S |
| 9 | 10 | 1 | 2 | Nasser, Mrs. Nicholas (Adele Achem) | female | 14.0 | 1 | 0 | 237736 | 30.0708 | NaN | C |
Try to add the "Age" column to our model:
- Fitting the pipeline will throw an error due to the presence of a missing value
- scikit-learn models don't accept data with missing values (except for Histogram-based Gradient Boosting Trees)
cols = ['Parch', 'Fare', 'Embarked', 'Sex', 'Name', 'Age']
X = df[cols]
X
| Parch | Fare | Embarked | Sex | Name | Age | |
|---|---|---|---|---|---|---|
| 0 | 0 | 7.2500 | S | male | Braund, Mr. Owen Harris | 22.0 |
| 1 | 0 | 71.2833 | C | female | Cumings, Mrs. John Bradley (Florence Briggs Th... | 38.0 |
| 2 | 0 | 7.9250 | S | female | Heikkinen, Miss. Laina | 26.0 |
| 3 | 0 | 53.1000 | S | female | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 35.0 |
| 4 | 0 | 8.0500 | S | male | Allen, Mr. William Henry | 35.0 |
| 5 | 0 | 8.4583 | Q | male | Moran, Mr. James | NaN |
| 6 | 0 | 51.8625 | S | male | McCarthy, Mr. Timothy J | 54.0 |
| 7 | 1 | 21.0750 | S | male | Palsson, Master. Gosta Leonard | 2.0 |
| 8 | 2 | 11.1333 | S | female | Johnson, Mrs. Oscar W (Elisabeth Vilhelmina Berg) | 27.0 |
| 9 | 0 | 30.0708 | C | female | Nasser, Mrs. Nicholas (Adele Achem) | 14.0 |
# pipe.fit(X, y)
One option is to drop any rows from the DataFrame that have missing values:
- This can be a useful approach, but only if you know that the missingness is random and it only affects a small portion of your dataset
- If a lot of your rows have missing values, then this approach will throw away too much useful training data
X.dropna()
| Parch | Fare | Embarked | Sex | Name | Age | |
|---|---|---|---|---|---|---|
| 0 | 0 | 7.2500 | S | male | Braund, Mr. Owen Harris | 22.0 |
| 1 | 0 | 71.2833 | C | female | Cumings, Mrs. John Bradley (Florence Briggs Th... | 38.0 |
| 2 | 0 | 7.9250 | S | female | Heikkinen, Miss. Laina | 26.0 |
| 3 | 0 | 53.1000 | S | female | Futrelle, Mrs. Jacques Heath (Lily May Peel) | 35.0 |
| 4 | 0 | 8.0500 | S | male | Allen, Mr. William Henry | 35.0 |
| 6 | 0 | 51.8625 | S | male | McCarthy, Mr. Timothy J | 54.0 |
| 7 | 1 | 21.0750 | S | male | Palsson, Master. Gosta Leonard | 2.0 |
| 8 | 2 | 11.1333 | S | female | Johnson, Mrs. Oscar W (Elisabeth Vilhelmina Berg) | 27.0 |
| 9 | 0 | 30.0708 | C | female | Nasser, Mrs. Nicholas (Adele Achem) | 14.0 |
A second option is to drop any features that have missing values:
- However, you may be throwing away a useful feature
X.dropna(axis='columns')
| Parch | Fare | Embarked | Sex | Name | |
|---|---|---|---|---|---|
| 0 | 0 | 7.2500 | S | male | Braund, Mr. Owen Harris |
| 1 | 0 | 71.2833 | C | female | Cumings, Mrs. John Bradley (Florence Briggs Th... |
| 2 | 0 | 7.9250 | S | female | Heikkinen, Miss. Laina |
| 3 | 0 | 53.1000 | S | female | Futrelle, Mrs. Jacques Heath (Lily May Peel) |
| 4 | 0 | 8.0500 | S | male | Allen, Mr. William Henry |
| 5 | 0 | 8.4583 | Q | male | Moran, Mr. James |
| 6 | 0 | 51.8625 | S | male | McCarthy, Mr. Timothy J |
| 7 | 1 | 21.0750 | S | male | Palsson, Master. Gosta Leonard |
| 8 | 2 | 11.1333 | S | female | Johnson, Mrs. Oscar W (Elisabeth Vilhelmina Berg) |
| 9 | 0 | 30.0708 | C | female | Nasser, Mrs. Nicholas (Adele Achem) |
A third option is to impute missing values:
- Imputation means that you are filling in missing values based on what you know from the non-missing data
- Carefully consider the costs and benefits of imputation before proceeding, because you are making up data
Use SimpleImputer to perform the imputation:
- It requires 2-dimensional input (just like OneHotEncoder)
- By default, it fills missing values with the mean of the non-missing values
- It also supports other imputation strategies: median value, most frequent value, or a user-defined value
from sklearn.impute import SimpleImputer
imp = SimpleImputer()
imp.fit_transform(X[['Age']])
array([[22. ],
[38. ],
[26. ],
[35. ],
[35. ],
[28.11111111],
[54. ],
[ 2. ],
[27. ],
[14. ]])
Examine the statistics_ attribute (which was learned during the fit step) to see what value was imputed:
imp.statistics_
array([28.11111111])
Update the ColumnTransformer to include the SimpleImputer:
- Brackets are required around "Age" because SimpleImputer expects 2-dimensional input
- Reminder: Brackets are not allowed around "Name" because CountVectorizer expects 1-dimensional input
ct = make_column_transformer(
(ohe, ['Embarked', 'Sex']),
(vect, 'Name'),
(imp, ['Age']),
remainder='passthrough')
ct.fit_transform(X)
<10x48 sparse matrix of type '<class 'numpy.float64'>' with 88 stored elements in Compressed Sparse Row format>
Update the Pipeline to include the revised ColumnTransformer, and fit it on X and y:
pipe = make_pipeline(ct, logreg)
pipe.fit(X, y);
Examine the "named_steps" to confirm that the Pipeline looks correct:
pipe.named_steps
{'columntransformer': ColumnTransformer(n_jobs=None, remainder='passthrough', sparse_threshold=0.3,
transformer_weights=None,
transformers=[('onehotencoder',
OneHotEncoder(categories='auto', drop=None,
dtype=<class 'numpy.float64'>,
handle_unknown='error',
sparse=True),
['Embarked', 'Sex']),
('countvectorizer',
CountVectorizer(analyzer='word', binary=False,
decode_error='strict',
dtype=...
input='content',
lowercase=True, max_df=1.0,
max_features=None, min_df=1,
ngram_range=(1, 1),
preprocessor=None,
stop_words=None,
strip_accents=None,
token_pattern='(?u)\\b\\w\\w+\\b',
tokenizer=None,
vocabulary=None),
'Name'),
('simpleimputer',
SimpleImputer(add_indicator=False, copy=True,
fill_value=None,
missing_values=nan,
strategy='mean', verbose=0),
['Age'])],
verbose=False),
'logisticregression': LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, l1_ratio=None, max_iter=100,
multi_class='auto', n_jobs=None, penalty='l2',
random_state=1, solver='liblinear', tol=0.0001, verbose=0,
warm_start=False)}
Update X_new to use the same columns as X, and then make predictions:
X_new = df_new[cols]
pipe.predict(X_new)
array([0, 0, 0, 0, 1, 0, 1, 0, 1, 0])
What happened during the predict step?
- If X_new didn't have any missing values in "Age", then nothing gets imputed during prediction
- If X_new did have missing values in "Age", then the imputation value is the mean of "Age" in X (which was 28.11), not the mean of "Age" in X_new
- This is important because you are only allowed to learn from the training data, and then apply what you learned to both the training and testing data
- This is why we fit_transform on training data, and transform (only) on testing data
- During prediction, every row (in X_new) is considered independently and predictions are done one at a time
- Thus if you passed a single row to the predict method, it becomes obvious that scikit-learn has to look to the training data for the imputation value
When imputing missing values, you can also add "missingness" as a feature:
- Set "add_indicator=True" (new in version 0.21) to add a binary indicator matrix indicating the presence of missing values
- This is useful when the data is not missing at random, since there might be a relationship between "missingness" and the target
- Example: If "Age" is missing because older passengers declined to give their ages, and older passengers are more likely to have survived, then there is a relationship between "missing Age" and "Survived"
imp_indicator = SimpleImputer(add_indicator=True)
imp_indicator.fit_transform(X[['Age']])
array([[22. , 0. ],
[38. , 0. ],
[26. , 0. ],
[35. , 0. ],
[35. , 0. ],
[28.11111111, 1. ],
[54. , 0. ],
[ 2. , 0. ],
[27. , 0. ],
[14. , 0. ]])
There are also other imputers available in scikit-learn:
- IterativeImputer (new in version 0.21)
- KNNImputer (new in version 0.22)
These new imputers will produce more useful imputations than SimpleImputer in some (but not all) cases.
Part 6: Switching to the full dataset¶
Read the full datasets into df and df_new:
df = pd.read_csv('http://bit.ly/kaggletrain')
df.shape
(891, 12)
df_new = pd.read_csv('http://bit.ly/kaggletest')
df_new.shape
(418, 11)
Check for missing values in the full datasets:
- There are two new problems we'll have to handle that weren't present in our smaller datasets:
- Problem 1: "Embarked" has missing values in df
- Problem 2: "Fare" has missing values in df_new
df.isna().sum()
PassengerId 0 Survived 0 Pclass 0 Name 0 Sex 0 Age 177 SibSp 0 Parch 0 Ticket 0 Fare 0 Cabin 687 Embarked 2 dtype: int64
df_new.isna().sum()
PassengerId 0 Pclass 0 Name 0 Sex 0 Age 86 SibSp 0 Parch 0 Ticket 0 Fare 1 Cabin 327 Embarked 0 dtype: int64
Redefine X and y for the full dataset:
X = df[cols]
y = df['Survived']
fit_transform will error since "Embarked" contains missing values (problem 1):
ct = make_column_transformer(
(ohe, ['Embarked', 'Sex']),
(vect, 'Name'),
(imp, ['Age']),
remainder='passthrough')
# ct.fit_transform(X)
We'll solve problem 1 by imputing missing values for "Embarked" before one-hot encoding it.
First create a new imputer:
- For categorical features, you can impute the most frequent value or a user-defined value
- We'll impute a user-defined value of "missing" (a string):
- This essentially treats missing values as a fourth category, and it will become a fourth column during one-hot encoding
- This is similar (but not identical) to imputing the most frequent value and then adding a missing indicator
imp_constant = SimpleImputer(strategy='constant', fill_value='missing')
Next create a Pipeline of two transformers:
- Step 1 is imputation, and step 2 is one-hot encoding
- fit_transform on "Embarked" now outputs four columns (rather than three)
imp_ohe = make_pipeline(imp_constant, ohe)
imp_ohe.fit_transform(X[['Embarked']])
<891x4 sparse matrix of type '<class 'numpy.float64'>' with 891 stored elements in Compressed Sparse Row format>
This is what happens "under the hood" when you fit_transform the Pipeline:
ohe.fit_transform(imp_constant.fit_transform(X[['Embarked']]))
<891x4 sparse matrix of type '<class 'numpy.float64'>' with 891 stored elements in Compressed Sparse Row format>
Here are the rules for Pipelines:
- All Pipeline steps (other than the final step) must be a transformer, and the final step can be a model or a transformer
- Our larger Pipeline (called "pipe") ends in a model, and thus we use the fit and predict methods with it
- Our smaller Pipeline (called "imp_ohe") ends in a transformer, and thus we use the fit_transform and transform methods with it
Replace "ohe" with "imp_ohe" in the ColumnTransformer:
- You can use any transformer inside of a ColumnTransformer, and "imp_ohe" is eligible since it acts like a transformer
- It's fine to apply "imp_ohe" to "Sex" as well as "Embarked":
- There are no missing values in "Sex" so the imputation step won't affect it
ct = make_column_transformer(
(imp_ohe, ['Embarked', 'Sex']),
(vect, 'Name'),
(imp, ['Age']),
remainder='passthrough')
We have solved problem 1, so we can now fit_transform on X:
- The feature matrix is much wider than before because "Name" has a ton of unique words
ct.fit_transform(X)
<891x1518 sparse matrix of type '<class 'numpy.float64'>' with 7328 stored elements in Compressed Sparse Row format>
We'll solve problem 2 by imputing missing values for "Fare":
- Modify the ColumnTransformer to apply the "imp" transformer to "Fare"
- Remember that "Fare" only has missing values in X_new, but not in X:
- When the imputer is fit to X, it will learn the imputation value that will be applied to X_new during prediction
ct = make_column_transformer(
(imp_ohe, ['Embarked', 'Sex']),
(vect, 'Name'),
(imp, ['Age', 'Fare']),
remainder='passthrough')
fit_transform outputs the same number of columns as before, since "Fare" just moved from a passthrough column to a transformed column:
ct.fit_transform(X)
<891x1518 sparse matrix of type '<class 'numpy.float64'>' with 7328 stored elements in Compressed Sparse Row format>
Update the Pipeline to include the revised ColumnTransformer, and fit it on X and y:
pipe = make_pipeline(ct, logreg)
pipe.fit(X, y);
Update X_new to use the same columns as X, and then make predictions:
X_new = df_new[cols]
pipe.predict(X_new)
array([0, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 1, 1, 0, 0, 0, 1, 0, 1,
1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1,
1, 0, 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 1,
1, 0, 0, 1, 1, 0, 1, 0, 1, 1, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 1, 1,
1, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0,
0, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0,
1, 0, 1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1,
1, 0, 1, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 1,
0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0,
1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1,
1, 0, 0, 0, 1, 0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1,
0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 1, 0, 0, 0, 0, 1, 0, 1, 1, 1, 0,
0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 1,
0, 1, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0,
1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 0, 1, 1, 0,
0, 0, 1, 0, 1, 0, 0, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 0,
1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1,
0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 0, 1, 0, 0, 1])
Recap¶
This is all of the code that is necessary to recreate our workflow up to this point:
- You can copy/paste this code from http://bit.ly/complex-pipeline
- There are no calls to "fit_transform" or "transform" because all of that functionality is encapsulated by the Pipeline
import pandas as pd
from sklearn.impute import SimpleImputer
from sklearn.preprocessing import OneHotEncoder
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.linear_model import LogisticRegression
from sklearn.compose import make_column_transformer
from sklearn.pipeline import make_pipeline
cols = ['Parch', 'Fare', 'Embarked', 'Sex', 'Name', 'Age']
df = pd.read_csv('http://bit.ly/kaggletrain')
X = df[cols]
y = df['Survived']
df_new = pd.read_csv('http://bit.ly/kaggletest')
X_new = df_new[cols]
imp_constant = SimpleImputer(strategy='constant', fill_value='missing')
ohe = OneHotEncoder()
imp_ohe = make_pipeline(imp_constant, ohe)
vect = CountVectorizer()
imp = SimpleImputer()
ct = make_column_transformer(
(imp_ohe, ['Embarked', 'Sex']),
(vect, 'Name'),
(imp, ['Age', 'Fare']),
remainder='passthrough')
logreg = LogisticRegression(solver='liblinear', random_state=1)
pipe = make_pipeline(ct, logreg)
pipe.fit(X, y)
pipe.predict(X_new)
array([0, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 1, 1, 0, 0, 0, 1, 0, 1,
1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1,
1, 0, 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 1,
1, 0, 0, 1, 1, 0, 1, 0, 1, 1, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 1, 1,
1, 1, 1, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0,
0, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0,
0, 0, 0, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0,
1, 0, 1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1,
1, 0, 1, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 1,
0, 1, 1, 0, 1, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0,
1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1, 1, 1,
1, 0, 0, 0, 1, 0, 1, 1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1,
0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 1, 0, 0, 0, 0, 1, 0, 1, 1, 1, 0,
0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 0, 1,
0, 1, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 0, 0, 0,
1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 0, 1, 1, 0,
0, 0, 1, 0, 1, 0, 0, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 0,
1, 1, 1, 0, 0, 1, 0, 0, 1, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1,
0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 1, 1, 1, 1, 1, 0, 1, 0, 0, 1])
Comparing Pipeline and ColumnTransformer:
- ColumnTransformer pulls out subsets of columns and transforms them independently, and then stacks the results side-by-side
- Pipeline is a series of steps that occur in order, and the output of each step passes to the next step
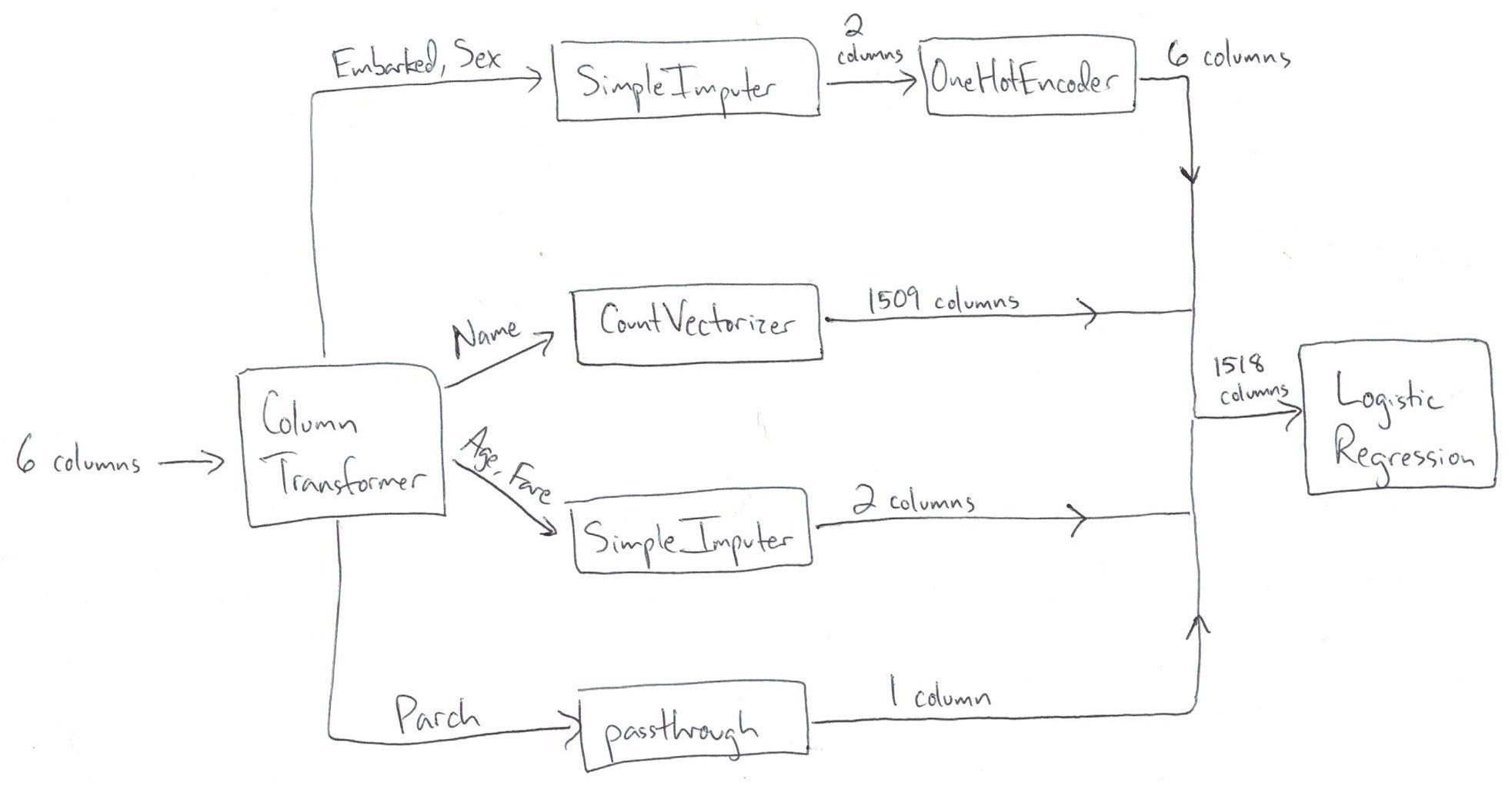
Why wouldn't we do all of the transformations in pandas, and just use scikit-learn for model building?
- CountVectorizer is a highly useful technique for encoding text data, and it can't be done using pandas
- Using both pandas and scikit-learn for transformations adds workflow complexity, especially if you have to combine a dense matrix (output by pandas) and a sparse matrix (output by CountVectorizer)
- One-hot encoding can be done using pandas, but you will probably add those columns to your DataFrame
- This makes the DataFrame larger and more difficult to navigate
- Missing value imputation can be done using pandas, but it will result in data leakage
What is data leakage?
- Inadvertently including knowledge from the testing data when training a model
Why is data leakage bad?
- Your model evaluation scores will be less reliable
- This may lead you to make bad decisions when tuning hyperparameters
- This will lead you to overestimate how well your model will perform on new data
- It's hard to know whether your scores will be off by a negligible amount or a huge amount
Why would missing value imputation in pandas cause data leakage?
- Your model evaluation procedure (such as cross-validation) is supposed to simulate the future, so that you can accurately estimate right now how well your model will perform on new data
- If you impute missing values on your whole dataset in pandas and then pass your dataset to scikit-learn, your model evaluation procedure will no longer be an accurate simulation of reality
- This is because the imputation values are based on your entire dataset, rather than just the training portion of your dataset
- Keep in mind that the "training portion" will change 5 times during 5-fold cross-validation, thus it's quite impractical to avoid data leakage if you use pandas for imputation
What other transformations in pandas will cause data leakage?
- Feature scaling
- One-hot encoding (unless there is a fixed set of categories)
- Any transformations which incorporate information about other rows when transforming a row
How does scikit-learn prevent data leakage?
- It has separate fit and transform steps, which allow you to base your data transformations on the training set only, and then apply those transformations to both the training set and the testing set
- Pipeline's fit and predict methods ensure that fit_transform and transform are called at the appropriate times
- cross_val_score and GridSearchCV split the data prior to performing data transformations
Part 7: Evaluating and tuning a Pipeline¶
We can use cross_val_score on the entire Pipeline to estimate its classification accuracy:
- Cross-validation is a useful tool now that we're using the full dataset
- We're using 5 folds because it has been shown to be a reasonable default choice
- cross_val_score performs the data transformations (specified in the ColumnTransformer) after each of the 5 data splits in order to prevent data leakage
- If it performed the data transformations before the data splits, that would have resulted in data leakage
from sklearn.model_selection import cross_val_score
cross_val_score(pipe, X, y, cv=5, scoring='accuracy').mean()
0.8114619295712762
Our next step is to tune the hyperparameters for both the model and the transformers:
- We have been using the default hyperparameters for most objects
- "Hyperparameters" are values you set, whereas "parameters" are values learned by the estimator during the fitting process
- Hyperparameter tuning is likely to result in a more accurate model
We'll use GridSearchCV for hyperparameter tuning:
- You define what values you want to try for each hyperparameter, and it cross-validates every possible combination of those values
- You have to tune hyperparameters together, since the best performing combination might be when none of them are at their default values
- Being able to tune the transformers simultaneous to the model is yet another benefit of doing transformations in scikit-learn rather than pandas
Because we're tuning a Pipeline, we need to know the step names from named_steps:
pipe.named_steps.keys()
dict_keys(['columntransformer', 'logisticregression'])
Specify the hyperparameters and values to try in a dictionary:
- Create an empty dictionary called params
- For our logistic regression model, we will tune:
- penalty: type of regularization (default is 'l2')
- C: amount of regularization (default is 1.0)
- Choosing which hyperparameters to tune and what values to try requires both research and experience
- The dictionary key is the step name, followed by 2 underscores, followed by the hyperparameter name
- The dictionary value is the list of values you want to try for that hyperparameter
params = {}
params['logisticregression__penalty'] = ['l1', 'l2']
params['logisticregression__C'] = [0.1, 1, 10]
params
{'logisticregression__penalty': ['l1', 'l2'],
'logisticregression__C': [0.1, 1, 10]}
Set up the grid search:
- Creating a GridSearchCV instance is similar to cross_val_score, except that you don't pass X and y but you do pass params
- Fitting the GridSearchCV object performs the grid search
from sklearn.model_selection import GridSearchCV
grid = GridSearchCV(pipe, params, cv=5, scoring='accuracy')
grid.fit(X, y);
Convert the results of the grid search into a DataFrame:
- 6 rows means that it ran cross-validation 6 times, which is every possible combination of C (3 values) and penalty (2 values)
results = pd.DataFrame(grid.cv_results_)
results
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_logisticregression__C | param_logisticregression__penalty | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0.013179 | 0.001090 | 0.006131 | 0.001403 | 0.1 | l1 | {'logisticregression__C': 0.1, 'logisticregres... | 0.787709 | 0.803371 | 0.769663 | 0.758427 | 0.797753 | 0.783385 | 0.016946 | 6 |
| 1 | 0.012467 | 0.000274 | 0.004867 | 0.000117 | 0.1 | l2 | {'logisticregression__C': 0.1, 'logisticregres... | 0.798883 | 0.803371 | 0.764045 | 0.775281 | 0.803371 | 0.788990 | 0.016258 | 5 |
| 2 | 0.013442 | 0.000392 | 0.004720 | 0.000045 | 1 | l1 | {'logisticregression__C': 1, 'logisticregressi... | 0.815642 | 0.820225 | 0.797753 | 0.792135 | 0.848315 | 0.814814 | 0.019787 | 2 |
| 3 | 0.012881 | 0.000346 | 0.004768 | 0.000058 | 1 | l2 | {'logisticregression__C': 1, 'logisticregressi... | 0.798883 | 0.825843 | 0.803371 | 0.786517 | 0.842697 | 0.811462 | 0.020141 | 3 |
| 4 | 0.018128 | 0.002229 | 0.004792 | 0.000173 | 10 | l1 | {'logisticregression__C': 10, 'logisticregress... | 0.821229 | 0.814607 | 0.814607 | 0.792135 | 0.848315 | 0.818178 | 0.018007 | 1 |
| 5 | 0.013615 | 0.000414 | 0.004737 | 0.000087 | 10 | l2 | {'logisticregression__C': 10, 'logisticregress... | 0.782123 | 0.803371 | 0.808989 | 0.797753 | 0.853933 | 0.809234 | 0.024080 | 4 |
Sort the DataFrame by "rank_test_score":
- Our column of interest is "mean_test_score"
- Best result was C=10 and penalty='l1', neither of which was the default
results.sort_values('rank_test_score')
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_logisticregression__C | param_logisticregression__penalty | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 4 | 0.018128 | 0.002229 | 0.004792 | 0.000173 | 10 | l1 | {'logisticregression__C': 10, 'logisticregress... | 0.821229 | 0.814607 | 0.814607 | 0.792135 | 0.848315 | 0.818178 | 0.018007 | 1 |
| 2 | 0.013442 | 0.000392 | 0.004720 | 0.000045 | 1 | l1 | {'logisticregression__C': 1, 'logisticregressi... | 0.815642 | 0.820225 | 0.797753 | 0.792135 | 0.848315 | 0.814814 | 0.019787 | 2 |
| 3 | 0.012881 | 0.000346 | 0.004768 | 0.000058 | 1 | l2 | {'logisticregression__C': 1, 'logisticregressi... | 0.798883 | 0.825843 | 0.803371 | 0.786517 | 0.842697 | 0.811462 | 0.020141 | 3 |
| 5 | 0.013615 | 0.000414 | 0.004737 | 0.000087 | 10 | l2 | {'logisticregression__C': 10, 'logisticregress... | 0.782123 | 0.803371 | 0.808989 | 0.797753 | 0.853933 | 0.809234 | 0.024080 | 4 |
| 1 | 0.012467 | 0.000274 | 0.004867 | 0.000117 | 0.1 | l2 | {'logisticregression__C': 0.1, 'logisticregres... | 0.798883 | 0.803371 | 0.764045 | 0.775281 | 0.803371 | 0.788990 | 0.016258 | 5 |
| 0 | 0.013179 | 0.001090 | 0.006131 | 0.001403 | 0.1 | l1 | {'logisticregression__C': 0.1, 'logisticregres... | 0.787709 | 0.803371 | 0.769663 | 0.758427 | 0.797753 | 0.783385 | 0.016946 | 6 |
In order to tune the transformers, we need to know their names:
pipe.named_steps.columntransformer.named_transformers_
{'pipeline': Pipeline(memory=None,
steps=[('simpleimputer',
SimpleImputer(add_indicator=False, copy=True,
fill_value='missing', missing_values=nan,
strategy='constant', verbose=0)),
('onehotencoder',
OneHotEncoder(categories='auto', drop=None,
dtype=<class 'numpy.float64'>,
handle_unknown='error', sparse=True))],
verbose=False),
'countvectorizer': CountVectorizer(analyzer='word', binary=False, decode_error='strict',
dtype=<class 'numpy.int64'>, encoding='utf-8', input='content',
lowercase=True, max_df=1.0, max_features=None, min_df=1,
ngram_range=(1, 1), preprocessor=None, stop_words=None,
strip_accents=None, token_pattern='(?u)\\b\\w\\w+\\b',
tokenizer=None, vocabulary=None),
'simpleimputer': SimpleImputer(add_indicator=False, copy=True, fill_value=None,
missing_values=nan, strategy='mean', verbose=0),
'remainder': 'passthrough'}
Tune the "drop" hyperparameter of OneHotEncoder by adding it to the params dictionary:
- Pipeline step: "columntransformer"
- First transformer: "pipeline"
- Second step of the inner pipeline: "onehotencoder"
- Hyperparameter: "drop"
- Separate each of these components by 2 underscores
Try the values None and 'first':
- None is the default
- 'first' means drop the first level of each feature after encoding (new in version 0.21)
params['columntransformer__pipeline__onehotencoder__drop'] = [None, 'first']
Tune the "ngram_range" hyperparameter of CountVectorizer:
- Pipeline step: "columntransformer"
- Second transformer: "countvectorizer"
- Hyperparameter: "ngram_range" (note the single underscore)
Try the values (1, 1) and (1, 2):
- (1, 1) is the default, which creates a single feature from each word
- (1, 2) creates features from both single words and word pairs
params['columntransformer__countvectorizer__ngram_range'] = [(1, 1), (1, 2)]
Tune the "add_indicator" hyperparameter of SimpleImputer:
- Pipeline step: "columntransformer"
- Third transformer: "simpleimputer"
- Hyperparameter: "add_indicator" (note the single underscore)
Try the values False and True:
- False is the default
- True means add a binary indicator matrix (new in version 0.21)
params['columntransformer__simpleimputer__add_indicator'] = [False, True]
Examine the params dictionary for any typos:
params
{'logisticregression__penalty': ['l1', 'l2'],
'logisticregression__C': [0.1, 1, 10],
'columntransformer__pipeline__onehotencoder__drop': [None, 'first'],
'columntransformer__countvectorizer__ngram_range': [(1, 1), (1, 2)],
'columntransformer__simpleimputer__add_indicator': [False, True]}
Perform the grid search again:
- There are 48 combinations to try, so it takes 8 times longer than the previous search
grid = GridSearchCV(pipe, params, cv=5, scoring='accuracy')
grid.fit(X, y);
Sort and review the search results:
- Accuracy of the best model is an improvement over the previous grid search
- It's hard to pick out trends for each hyperparameter because many of them affect one another
results = pd.DataFrame(grid.cv_results_)
results.sort_values('rank_test_score')
| mean_fit_time | std_fit_time | mean_score_time | std_score_time | param_columntransformer__countvectorizer__ngram_range | param_columntransformer__pipeline__onehotencoder__drop | param_columntransformer__simpleimputer__add_indicator | param_logisticregression__C | param_logisticregression__penalty | params | split0_test_score | split1_test_score | split2_test_score | split3_test_score | split4_test_score | mean_test_score | std_test_score | rank_test_score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 28 | 0.023061 | 0.002269 | 0.005146 | 0.000024 | (1, 2) | None | False | 10 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.860335 | 0.820225 | 0.820225 | 0.786517 | 0.859551 | 0.829370 | 0.027833 | 1 |
| 46 | 0.029656 | 0.003894 | 0.005461 | 0.000197 | (1, 2) | first | True | 10 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.849162 | 0.831461 | 0.820225 | 0.786517 | 0.853933 | 0.828259 | 0.024138 | 2 |
| 40 | 0.030675 | 0.002119 | 0.005186 | 0.000081 | (1, 2) | first | False | 10 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.854749 | 0.825843 | 0.814607 | 0.786517 | 0.848315 | 0.826006 | 0.024549 | 3 |
| 34 | 0.023133 | 0.001805 | 0.005422 | 0.000201 | (1, 2) | None | True | 10 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.849162 | 0.820225 | 0.820225 | 0.780899 | 0.853933 | 0.824889 | 0.026120 | 4 |
| 10 | 0.020138 | 0.002229 | 0.005470 | 0.000890 | (1, 1) | None | True | 10 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.826816 | 0.814607 | 0.820225 | 0.780899 | 0.853933 | 0.819296 | 0.023467 | 5 |
| 22 | 0.021249 | 0.001699 | 0.004953 | 0.000112 | (1, 1) | first | True | 10 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.821229 | 0.803371 | 0.825843 | 0.780899 | 0.859551 | 0.818178 | 0.026034 | 6 |
| 4 | 0.018240 | 0.001837 | 0.004744 | 0.000091 | (1, 1) | None | False | 10 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.821229 | 0.814607 | 0.814607 | 0.792135 | 0.848315 | 0.818178 | 0.018007 | 6 |
| 20 | 0.014477 | 0.001047 | 0.005164 | 0.000368 | (1, 1) | first | True | 1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.810056 | 0.820225 | 0.797753 | 0.792135 | 0.853933 | 0.814820 | 0.021852 | 8 |
| 2 | 0.013728 | 0.000415 | 0.004839 | 0.000075 | (1, 1) | None | False | 1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.815642 | 0.820225 | 0.797753 | 0.792135 | 0.848315 | 0.814814 | 0.019787 | 9 |
| 16 | 0.021138 | 0.001391 | 0.004800 | 0.000139 | (1, 1) | first | False | 10 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.821229 | 0.803371 | 0.814607 | 0.780899 | 0.853933 | 0.814808 | 0.023886 | 10 |
| 44 | 0.018747 | 0.001117 | 0.005938 | 0.000527 | (1, 2) | first | True | 1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.804469 | 0.820225 | 0.797753 | 0.792135 | 0.853933 | 0.813703 | 0.022207 | 11 |
| 47 | 0.018135 | 0.000447 | 0.005382 | 0.000114 | (1, 2) | first | True | 10 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.787709 | 0.820225 | 0.820225 | 0.780899 | 0.853933 | 0.812598 | 0.026265 | 12 |
| 8 | 0.013765 | 0.000456 | 0.004881 | 0.000127 | (1, 1) | None | True | 1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.804469 | 0.820225 | 0.786517 | 0.792135 | 0.859551 | 0.812579 | 0.026183 | 13 |
| 14 | 0.013688 | 0.000971 | 0.004796 | 0.000181 | (1, 1) | first | False | 1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.804469 | 0.820225 | 0.797753 | 0.792135 | 0.848315 | 0.812579 | 0.020194 | 14 |
| 38 | 0.017634 | 0.000525 | 0.005225 | 0.000081 | (1, 2) | first | False | 1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.804469 | 0.820225 | 0.797753 | 0.792135 | 0.848315 | 0.812579 | 0.020194 | 14 |
| 11 | 0.014208 | 0.000597 | 0.005329 | 0.000715 | (1, 1) | None | True | 10 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.782123 | 0.803371 | 0.808989 | 0.792135 | 0.870787 | 0.811481 | 0.031065 | 16 |
| 21 | 0.013204 | 0.000708 | 0.004958 | 0.000345 | (1, 1) | first | True | 1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.793296 | 0.820225 | 0.803371 | 0.786517 | 0.853933 | 0.811468 | 0.024076 | 17 |
| 3 | 0.013157 | 0.000342 | 0.004966 | 0.000461 | (1, 1) | None | False | 1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.798883 | 0.825843 | 0.803371 | 0.786517 | 0.842697 | 0.811462 | 0.020141 | 18 |
| 26 | 0.017373 | 0.000133 | 0.005122 | 0.000050 | (1, 2) | None | False | 1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.810056 | 0.820225 | 0.786517 | 0.792135 | 0.848315 | 0.811449 | 0.022058 | 19 |
| 23 | 0.013665 | 0.000259 | 0.004913 | 0.000128 | (1, 1) | first | True | 10 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.776536 | 0.803371 | 0.808989 | 0.792135 | 0.870787 | 0.810363 | 0.032182 | 20 |
| 9 | 0.012987 | 0.000245 | 0.004787 | 0.000074 | (1, 1) | None | True | 1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.793296 | 0.825843 | 0.797753 | 0.786517 | 0.848315 | 0.810345 | 0.023233 | 21 |
| 15 | 0.012510 | 0.000078 | 0.004724 | 0.000065 | (1, 1) | first | False | 1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.804469 | 0.820225 | 0.803371 | 0.786517 | 0.837079 | 0.810332 | 0.017107 | 22 |
| 32 | 0.017513 | 0.000521 | 0.005245 | 0.000034 | (1, 2) | None | True | 1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.804469 | 0.820225 | 0.780899 | 0.792135 | 0.853933 | 0.810332 | 0.025419 | 22 |
| 17 | 0.013241 | 0.000162 | 0.004707 | 0.000095 | (1, 1) | first | False | 10 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.782123 | 0.803371 | 0.808989 | 0.797753 | 0.853933 | 0.809234 | 0.024080 | 24 |
| 35 | 0.018231 | 0.000590 | 0.005368 | 0.000091 | (1, 2) | None | True | 10 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.782123 | 0.820225 | 0.814607 | 0.780899 | 0.848315 | 0.809234 | 0.025357 | 24 |
| 5 | 0.013436 | 0.000172 | 0.004653 | 0.000031 | (1, 1) | None | False | 10 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.782123 | 0.803371 | 0.808989 | 0.797753 | 0.853933 | 0.809234 | 0.024080 | 24 |
| 29 | 0.023017 | 0.011150 | 0.005115 | 0.000026 | (1, 2) | None | False | 10 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.787709 | 0.814607 | 0.820225 | 0.780899 | 0.837079 | 0.808104 | 0.020904 | 27 |
| 45 | 0.017329 | 0.000598 | 0.005484 | 0.000115 | (1, 2) | first | True | 1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.793296 | 0.814607 | 0.797753 | 0.786517 | 0.848315 | 0.808097 | 0.022143 | 28 |
| 41 | 0.017454 | 0.000328 | 0.005192 | 0.000138 | (1, 2) | first | False | 10 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.787709 | 0.814607 | 0.820225 | 0.780899 | 0.831461 | 0.806980 | 0.019414 | 29 |
| 39 | 0.016762 | 0.000371 | 0.005216 | 0.000141 | (1, 2) | first | False | 1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.798883 | 0.808989 | 0.797753 | 0.786517 | 0.837079 | 0.805844 | 0.017164 | 30 |
| 27 | 0.016690 | 0.000149 | 0.005101 | 0.000031 | (1, 2) | None | False | 1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.798883 | 0.814607 | 0.792135 | 0.786517 | 0.837079 | 0.805844 | 0.018234 | 30 |
| 33 | 0.016940 | 0.000164 | 0.005267 | 0.000074 | (1, 2) | None | True | 1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.782123 | 0.814607 | 0.792135 | 0.786517 | 0.848315 | 0.804739 | 0.024489 | 32 |
| 31 | 0.016125 | 0.000202 | 0.005330 | 0.000113 | (1, 2) | None | True | 0.1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.798883 | 0.803371 | 0.769663 | 0.786517 | 0.814607 | 0.794608 | 0.015380 | 33 |
| 7 | 0.012880 | 0.001058 | 0.005017 | 0.000315 | (1, 1) | None | True | 0.1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.798883 | 0.803371 | 0.764045 | 0.786517 | 0.814607 | 0.793484 | 0.017253 | 34 |
| 19 | 0.012406 | 0.000379 | 0.004833 | 0.000086 | (1, 1) | first | True | 0.1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.793296 | 0.803371 | 0.764045 | 0.780899 | 0.814607 | 0.791243 | 0.017572 | 35 |
| 43 | 0.016018 | 0.000072 | 0.005258 | 0.000042 | (1, 2) | first | True | 0.1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.798883 | 0.797753 | 0.764045 | 0.780899 | 0.808989 | 0.790114 | 0.015849 | 36 |
| 37 | 0.016297 | 0.001262 | 0.005391 | 0.000457 | (1, 2) | first | False | 0.1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.787709 | 0.803371 | 0.764045 | 0.780899 | 0.808989 | 0.789003 | 0.016100 | 37 |
| 25 | 0.015791 | 0.000137 | 0.005094 | 0.000033 | (1, 2) | None | False | 0.1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.793296 | 0.803371 | 0.764045 | 0.775281 | 0.808989 | 0.788996 | 0.016944 | 38 |
| 1 | 0.012500 | 0.000984 | 0.004949 | 0.000352 | (1, 1) | None | False | 0.1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.798883 | 0.803371 | 0.764045 | 0.775281 | 0.803371 | 0.788990 | 0.016258 | 39 |
| 13 | 0.011891 | 0.000116 | 0.004815 | 0.000203 | (1, 1) | first | False | 0.1 | l2 | {'columntransformer__countvectorizer__ngram_ra... | 0.782123 | 0.803371 | 0.764045 | 0.780899 | 0.808989 | 0.787885 | 0.016343 | 40 |
| 0 | 0.014173 | 0.001353 | 0.005162 | 0.000298 | (1, 1) | None | False | 0.1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.787709 | 0.803371 | 0.769663 | 0.758427 | 0.797753 | 0.783385 | 0.016946 | 41 |
| 24 | 0.015586 | 0.000120 | 0.005175 | 0.000132 | (1, 2) | None | False | 0.1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.787709 | 0.803371 | 0.769663 | 0.758427 | 0.797753 | 0.783385 | 0.016946 | 41 |
| 6 | 0.012031 | 0.000139 | 0.004970 | 0.000330 | (1, 1) | None | True | 0.1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.787709 | 0.803371 | 0.769663 | 0.758427 | 0.797753 | 0.783385 | 0.016946 | 41 |
| 30 | 0.016099 | 0.000485 | 0.005302 | 0.000050 | (1, 2) | None | True | 0.1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.782123 | 0.803371 | 0.769663 | 0.758427 | 0.797753 | 0.782267 | 0.016807 | 44 |
| 36 | 0.015749 | 0.000290 | 0.005128 | 0.000038 | (1, 2) | first | False | 0.1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.770950 | 0.797753 | 0.769663 | 0.758427 | 0.792135 | 0.777785 | 0.014779 | 45 |
| 42 | 0.016111 | 0.000190 | 0.005278 | 0.000020 | (1, 2) | first | True | 0.1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.770950 | 0.797753 | 0.769663 | 0.758427 | 0.792135 | 0.777785 | 0.014779 | 45 |
| 12 | 0.012544 | 0.000763 | 0.004768 | 0.000070 | (1, 1) | first | False | 0.1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.770950 | 0.797753 | 0.769663 | 0.758427 | 0.792135 | 0.777785 | 0.014779 | 45 |
| 18 | 0.012307 | 0.000567 | 0.004854 | 0.000064 | (1, 1) | first | True | 0.1 | l1 | {'columntransformer__countvectorizer__ngram_ra... | 0.770950 | 0.797753 | 0.769663 | 0.758427 | 0.792135 | 0.777785 | 0.014779 | 45 |
Access the single best score and best set of hyperparameters:
- Two of the hyperparameters used the default values (drop, add_indicator)
- Three of the hyperparameters did not use the default values (ngram_range, C, penalty)
grid.best_score_
0.8293704098926622
grid.best_params_
{'columntransformer__countvectorizer__ngram_range': (1, 2),
'columntransformer__pipeline__onehotencoder__drop': None,
'columntransformer__simpleimputer__add_indicator': False,
'logisticregression__C': 10,
'logisticregression__penalty': 'l1'}
You can use the GridSearchCV object to make predictions:
- It automatically refits the Pipeline on all of the data (X and y) with the best set of hyperparameters
grid.predict(X_new)
array([0, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 1, 0, 1, 1, 0, 0, 1, 1, 0, 1,
1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1,
1, 0, 0, 0, 1, 1, 0, 0, 1, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 1,
1, 0, 0, 1, 1, 0, 1, 0, 1, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 1, 1,
1, 1, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 1, 0, 0, 0, 0, 0,
0, 1, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0,
0, 0, 0, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 1,
1, 0, 1, 1, 0, 1, 1, 1, 1, 0, 0, 1, 0, 0, 1, 0, 0, 0, 0, 0, 0, 1,
1, 0, 1, 1, 0, 0, 1, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 1, 0, 1, 1,
0, 1, 1, 1, 0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 1, 0, 0, 1, 0, 1, 0,
1, 0, 1, 0, 1, 1, 0, 1, 0, 0, 0, 1, 0, 0, 1, 0, 0, 0, 1, 1, 1, 1,
1, 0, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 1,
0, 0, 0, 0, 1, 0, 0, 0, 1, 1, 0, 1, 0, 0, 0, 0, 1, 1, 1, 0, 1, 0,
0, 0, 0, 0, 0, 1, 0, 0, 0, 0, 1, 1, 0, 0, 0, 0, 0, 0, 1, 1, 0, 1,
0, 1, 0, 0, 0, 1, 1, 1, 1, 0, 0, 0, 0, 0, 0, 0, 1, 1, 1, 0, 0, 0,
1, 0, 0, 1, 0, 0, 0, 0, 0, 1, 0, 0, 0, 1, 0, 1, 0, 1, 0, 1, 1, 0,
0, 0, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 1, 0, 0, 1, 1, 0, 0, 1, 0, 0,
1, 1, 0, 0, 0, 1, 0, 0, 1, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 0, 0, 1,
0, 1, 0, 0, 1, 0, 1, 0, 0, 0, 0, 0, 1, 1, 1, 1, 1, 0, 1, 0, 0, 1])