Table of Contents¶
PEB Belgrade - Bash workshop¶
Giovanni M. Dall'Olio, GlaxoSmithKline, 12/09/2016. All materials available here: https://dalloliogm.github.io/
_______________
/ Welcome to \
\ PEB Belgrade! /
---------------
\ ^__^
\ (oo)\_______
(__)\ )\/\
||----w |
|| ||
Welcome to Belgrade!
In this workshop we will review some basic Unix command, as well as bash usage.
If you attended the Programming for Evolutionary Biology course in Leipzig, this will be a refreshener. I've hidden some secrets in the exercises, so you will not get bored :-)
If you are new to bash, this will be a short introduction. Press Space or Down do continue.
Definitions: The Unix Philosophy¶
Unix is the name of an operating system created in the '80s, which became popular for introducing a novel approach to computing.
The Unix philosophy can be summarised as:
- Make each program do one thing well.
- Expect the output of every program to become the input to another, as yet unknown, program.
Press Space to continue.
You will see how each Unix tool is specialized on a single task, and how the piping system allows to combine these tool together.
These principles can be useful to any person wishing to learn programming. You may use the same approach when learning programming, starting writing small programs and functions, and combining them together in bigger pipelines.
More definitions¶
Linux:
A "descendant" of Unix, e.g. an operating system based on Unix that can run on modern computers
Terminal:
A software that allows to input commands to the computer, by typing them rather than point-and-click
Bash:
A command-line interpreter, e.g. a software that interprets the commands given from the terminal, and execute them.
How to follow this workshop¶
Getting a Terminal application¶
All the exercises will be done in a Terminal.
During the conference we may have also time for a "Linux Install Party", to get Linux into some of your laptops. However there are ways to access a bash terminal without installing Linux first.
Press space or the down key to see what to install or launch.
Windows¶
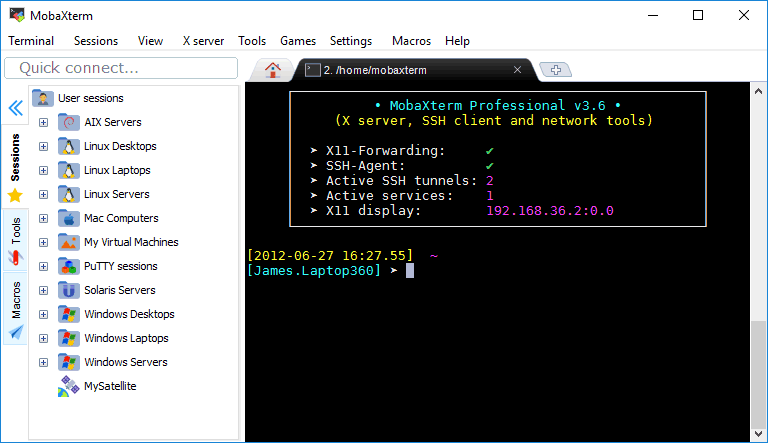
For Windows users, we will use a terminal emulator software called MobaXTerm: http://mobaxterm.mobatek.net/
The Home Edition is free and contains all the features we will need for the workshop:
To install new software, use (e.g. make):
apt-cyg install make
Linux¶
Congratulations on having Linux installed! You can use your favorite terminal app (e.g. gnome-terminal or konsole)
Getting the workshop materials¶
Now that we have a terminal application ready, let's download all the course materials.
Open the terminal, and type the following commands (omitting the "$:"):
$: wget https://github.com/dalloliogm/belgrade_unix_intro/archive/master.zip
$: unzip master.zip
Explanation:
- the wget command downloads a .zip file containing all the materials
- the unzip command uncompresses the .zip file, creating a new folder in your home area.
If you get "command not found"¶
Download https://github.com/dalloliogm/belgrade_unix_intro/archive/master.zip
Open Cygwin
cd /cygdrive/c/Documents\ and\ Settings/ (your name)
# Expected output
$: wget https://github.com/dalloliogm/belgrade_unix_intro/archive/master.zip
--2016-08-26 09:55:53-- https://github.com/dalloliogm/belgrade_unix_intro/archive/master.zip
Resolving github.com... 192.30.253.113
Connecting to github.com|192.30.253.113|:443... connected.
HTTP request sent, awaiting response... 302 Found
Location: https://codeload.github.com/dalloliogm/belgrade_unix_intro/zip/master [following]
--2016-08-26 09:55:54-- https://codeload.github.com/dalloliogm/belgrade_unix_intro/zip/master
Resolving codeload.github.com... 192.30.253.120
Connecting to codeload.github.com|192.30.253.120|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 129112 (126K) [application/zip]
Saving to: `master.zip.1'
100%[========================================================================================================================================================================>] 129,112 617K/s in 0.2s
2016-08-26 09:55:54 (617 KB/s) - `master.zip' saved [129112/129112]
$: unzip master.zip
Archive: master.zip
creating: belgrade_unix_intro-master/
inflating: belgrade_unix_intro-master/PEB Bash Workshop.ipynb
inflating: belgrade_unix_intro-master/README.md
creating: belgrade_unix_intro-master/data/
creating: belgrade_unix_intro-master/data/part1_grep/
inflating: belgrade_unix_intro-master/data/part1_grep/file1.txt
inflating: belgrade_unix_intro-master/data/part1_grep/file2.txt
creating: belgrade_unix_intro-master/src/
creating: belgrade_unix_intro-master/src/data/
inflating: belgrade_unix_intro-master/src/data/README.rst
inflating: belgrade_unix_intro-master/src/generate_grep_exercise.py
Advanced: using git to get the materials¶
If the software git is installed, you can get the materials by the following:
git clone git@github.com:dalloliogm/belgrade_unix_intro.git
Basic Unix Commands: ls, cd¶
Let's have a look at the files we just downloaded.
We will use two basic Unix commands:
- ls list the number of files in the current directory
- cd allows to move to a different directory.
Typing ls will show all the files in the current directory. Among these you should see a folder called belgrade_unix_intro-master, created by the wget and unzip commands.
Let's move to this new directory, and list the files in it:
$: cd belgrade_unix_intro-master/
$: ls
This will show a list of files, including a file called start_here.txt, a README, a few folders (data/, src/), and some other files.
ls
data PEB Bash Workshop.slides.html src Makefile PEB Bioconductor workshop.ipynb start_here.txt PEB Bash Workshop.ipynb README.md
Press space or the down key to continue.
ls -l¶
You can use the -l option of ls to visualize more details:
# Contents of the PEB workshop directory
ls -l
total 160 drwxrwxr-x 4 gioby gioby 4096 Sep 8 19:02 data -rw-rw-r-- 1 gioby gioby 929 Sep 8 21:57 Makefile -rw-rw-r-- 1 gioby gioby 83026 Sep 8 22:04 PEB Bash Workshop.ipynb -rw-rw-r-- 1 gioby gioby 56603 Sep 8 19:20 PEB Bioconductor workshop.ipynb -rw-rw-r-- 1 gioby gioby 260 Sep 5 18:23 README.md drwxrwxr-x 3 gioby gioby 4096 Sep 8 21:38 src -rw-rw-r-- 1 gioby gioby 1877 Sep 5 18:23 start_here.txt
Accessing the contents of a file: head, cat, less¶
The new folder contains a file named start_here.txt, containing the first instructions for the workshop.
To access the contents of a file, we can use several Unix commands:
| command | description | example |
|---|---|---|
| head | print the first lines of the file | head start_here.txt |
| tail | print the last lines of the file | tail start_here.txt |
| cat | print the contents of the file to the screen | cat start_here.txt |
| less | allows to navigate contents of the file | less start_here.txt |
For the first exercise, type "head start_here.txt" and follow the instructions:
head start_here.txt
_________________________________________________
/ To navigate the contents of this file, type: \
| |
| less start_here.txt |
\ /
--------------------------------------------------
\ ^__^
\ (oo)\_______
(__)\ )\/\
||----w |
Searching patterns into file: grep¶
The instructions for the next exercises are stored in the data/exercise1_grep.txt file.
However, if you look at this file with head or less, you will see that its contents have no meaning!
head data/exercise1_grep.txt
PWapg3ZDqzNWF6v VocznrWXXLTi gIY7Tj0bVx pmslXrBubMQeoEJXrF0OfHpcpxwTlktHSCm spf5840ZpMkpg4tZvgd3z4dxLVLiXnfmtrNaGL9d BV04Lu18iLMugTwPHRRkLCADC8PKO8jXutZ zBTK9i8ya oe4IoxbCZhST4XvDe mrccT7cwYGAD 1SmSareQB5q8wNsAvaA79aqXlIpmBZgmUVR 4gr mwZcxIg6pQwgddsJa4giM7hzjp8lit49D7kH upYIZQr8MbyEk4CX Y7k0uMmW9kk1fNJDea DMj0BJp wJ8BF3xyd61euAWb4IjOv6paBlKGse3a buZnpSOJv9PWhQpQnuxmZosVdYFw6TZF3RG yxArpyFCKt5637qiASyfadyheMBAp4bccq5furIx EOgGCEnWGuJwLSmvoehnXBdlbqDS5YN f7k T016mr v0mzedsHTFReC3ZjqVuYXpPuTulu8F0Z3 pmr9l96nOUEVXckfdiidZUP6UvFNh4Doaqz B0zFnnWEFttxrUjyuHgU9U09wEt7HfHBP1MAstQb WgxYhtDn3swa5fsmYgtxQKjjbIZzuVszEdl qByK4hFg7JQowOAXW60EBXQYSDHFgUWHlJAGYnjO CoB6YKtvZPaS8H8BRdsuBwdqU3KRz O9oXk 3Ntf9b6jv7hZsjtfEcaIzMuakpsEjl6i7Mra4M3U MgWDXcpafKACEA0rUAro9DjHo4VgbBJ6tdj
This is because I've hidden all the instructions in the file, just to make things more interesting!
Press space or the down key to see how to continue.
I've hidden the instructions for the first exercise in the lines containing the word "start" somewhere.
To find them, type:
$: grep start data/exercise1_grep
grep start data/exercise1_grep.txt
NjIM startesMsuqZNWhowFxuSFX4IaLymKGYdef McQYo6 umUY816rvtSGjAl start DdBaWfylxrZ ______________________________ Ewu1xLvv7OrXNWu4otWYoF gdV4U3istartdzYlJ / Congrats! \ kDDgWqtLBgY85 PQm8p1ajcAEzbQdb start rMv | You've used grep correctly, | kCVqk6sGesHvBp6 pNLzStgdhKu start 9YQQNI \ and found a cow. / tLMfr startUY36ToJEfE4uqIAQ3JboyoBOFyL8s ------------------------------ bWKJdeuL startI4xvVOZxwyC7oMKHaoG5ePF4k \ ^__^ fThKk5wk startAo6IzNddHcxuj93oFRam0mneoF \ (oo)\_______ awstartyw5tH3FzetzVxhw8c VrV7Uyis 5q8Yvj (__)\ )\/\ QgF4gHcEbAz start 9ZbzG90fUafm64BIlTEIIr ||----w | XKWA start6DpuNhYTXTcmo0UtCGa4SUo4JvnwvD || || ZA0BrMOyH99y7VY97lkomNXHUJUv8MWg start R YgOF6ahX0hEhMf startTZd 1 wDtbgoa86I8Atk vln0CxxjrcgeeQ5EtPdG0Spx7startIAT35hzj 6 xglkzByTDeiIKyoZbCQbO4brstart rb39 ExT9F The command grep allows to search 5start7zPVWugW3vb9 mYBIxsuIVxhHUdIxiTgFZ for a pattern in a text file. CLcSSkWNF0tHLOluZr43qptA startxojHnAwbmJ Fp start ZwtvOeMtDld9oahg9rdmBvKtIjPqXFQ It will print all the matching VOsyrwG4UOEsdfYLOfGFGKZWEvtJse start 9Ra lines to the screen. Sv81TKcZ8Fx1lb7xPZVMxW4ODNoKg8p7IHZstart GlbVstartpQQ5eQDweIn0VAGC8bQLbQ0Dzw4Ggvt ================= kziTL5jTistart pijnXXmWRApPCC 19SUNHN8n7 Next Exercise v768DQ0dRCix6 startc0me0SF qIsYfeC704lam ================= vebdjvHTdstart RxBxhJayFkmRXqyOvqg5khG4O QorxdcpNP1utzBstart6WpDOX4YzyIFpkZEalKW4 In the next exercise we will see GipBzz4Ul5sj3hVmVkQvPg startz9v6AF91EirG how to access grep's documentation. CC09wNO65rwuCqUgi8Skg1NZ0SGR7WDUoVTstart fjT7Ag59 RuhusLFzU startGHFvKsYSp bnNsLG Grep the following word to continue: Zx6RINR3hk start667gnhTiLYLiB30MxX7irwVP _ _ T0aoAQpfbNkO8LkSzSLJkLVEaXNxzQ startVoL3 | | | | Nv0hZYvh0pHN0AlT BNstart C8pMzkIs7usQUWd | |__ ___ | | _ __ 5 start 9Rq5tBOFDxiExQrRlPgCXoWt43a3US | '_ \ / _ \| || '_ \ 46unsRj3c4ClXQvcoFPyE9cnRHDQOHFNNZstartc | | | || __/| || |_) | 40H3 startj6glBCFXqOhMH3BEdgBsPPQuBbOt6D |_| |_| \___||_|| .__/ Qam1yoNK3BCpwSyhRX8Wb3rA1Ustart djDiAHuT | | PFW startKxEzUChDZGSPQNj4gsTS5k1JMBvWuY |_| 1bS5w1uaq65startnVRWYkojLFMSkMjui8YYz1A5 g0 g8iyP startQqkz7F05ST C S73TpreeesnFm
Accessing grep documentation¶
To access the documentation of a command, we can use the man command.
Let's type the following:
$: man grep
This will allow to navigate the documentation for grep, in the same modality as with the less command. Use the arrows to scroll, and q to exit.
For the next exercise, you will have to identify two options in the man page, and use them to do a case-insensitive search for "ignorecase", and count the number of lines.
grep help data/exercise1_grep.txt
td4kwN6cV0kqU3qMkwYOHl9MqjTQ help MP6MrF GY help yEvNL3RuVQYqiumisBftk8irLzXwt61y The documentation for grep can MTEwAhelphQapljO9yUtucAiNpvZrKdbwc3KUcsu be accessed through man: NVD5n5HKKKz6GgDmyOGMlKSMTd help Rii BCjC ku1GNL6IpSHBvcGroqpHgbMUNCg3Yz3lhelpnOBy $: man grep XkHkwMIhelp hidg7uJURR6loj5IAwv9oyeIUmqT sGKar9AKY help VhNi3MlGzT3WjAQdpWbvtuWeb Scroll down to see all the jHLbw4whFT1BhelpDfqZqhjXRYPjF0y7pkM8g3z3 parameters for grep and their description. hZ1OQgKcsgZo help m4s64C8nSR5zM gU4fYObu 9jlkynOWhelpLTaeswR5UnouUc3Ipsd4OjVI5PFO Use / to search for text. k4pjhosSNRgJlr7kthelpAvkWOHszFMoP yPEbgT Press the q key to exit. DPdj3lg4P6 UtuibInPhelp ErdkiRKtYHTKDAJ 5Ru8b5help cCuwtAVpbxoqHtK70dT9vtw5NsZR8 Ohelp hL9xy8U77RDjkZsRX6WDZf8ywnBY83LiL5 8nhxNAlz3yHtfZFEBjwvnKPFB help qUV YkeQX ============== FU4nthelp RStwArEw6UGFM4O7kKlxItNqVfD8Jl Next exercise IcOj help P56z3xD7QRkE1admG5sNTulg7B38om ============== FSnpSHWkiELodvyTu Txhelp uAWnw9UTW0ZPITU iCu7cLxdU0vLMBohelp46htatY8jYJC6XXVNDHTF For the next exercise, you will need to open qLbjcYhelp cp8USrp6u51ainbnXsp DForAbOq3 grep's documentation and identify two options: LPTakIcUOWmROON8GPJ4szSpKqZn3c help k5jK graSN0 cI4H6Zl help hCxcK0ynPImVu0Mogdcw - the option for case-insensitive searches MSewEHXyuatdRzy9GSokR DaLKphelpfDDJd7u p - the option for counting r7k6b1c9XDZcWnxH syn9peY uNqhelpjKyOyg0T the number of matching lines, zi4Rycq58rmxjH zW1AhCWAO1shelpSyViqAbyAC instead of printing them to the screen. CNx6GsFSshelp iRQE6pdA0jJiStNjknOaoQPSD ial36NIIePB7P5 help tpJ6bnVvVv7gESXp1Apc Once you have identified these options, A9HSI nKdCcuDp8WGEFkbWE8gJsUAZatatIOhelp do a case-insensitive search on this file for the word erU3 7ppIkaPoqBFCFkFFYMohelp HxVST S9fFj "ignorecase", then count the number of lines. lwjWEVzMBJSZiRSXvJzQmePQPFKeL4OQdOhelp R 8P5kONdSaqg0tolHUGq8nN9brT7k help 6duGCw
# If we do a search for "ignorecase" without any option, we only get some of the lines.
# You can notice that the cow is not properly displayed :-)
grep ignorecase data/exercise1_grep.txt
p14PGGXignorecaseDoCCJ9sYiegaozfL6LXxDmf o6m1cg7CignorecaseUJbpjD laYkpG6gdBHbJIM aNqS0Tg4kVIeLlyDeYoBlalps0ignorecasew5dd Remember that, to continue with the exercise, bRe7rR0sM8mcf8W1woMoReyjignorecaseLtPrHA you need to do a case-insensitive search for the word erU3 7ppIkaPoqBFCFkFFYMohelp HxVST S9fFj "ignorecase", then count the number of lines. wignorecaseTt0lDGMb5KCFWEm4t8RmBNXtLvURX ||----w |
# The -i option allows to do a case-insensitive search.
# As you can see, some lines contain upper case characters:
grep -i ignorecase data/exercise1_grep.txt
p14PGGXignorecaseDoCCJ9sYiegaozfL6LXxDmf o6m1cg7CignorecaseUJbpjD laYkpG6gdBHbJIM aNqS0Tg4kVIeLlyDeYoBlalps0ignorecasew5dd Remember that, to continue with the exercise, bRe7rR0sM8mcf8W1woMoReyjignorecaseLtPrHA you need to do a case-insensitive search for the word erU3 7ppIkaPoqBFCFkFFYMohelp HxVST S9fFj "ignorecase", then count the number of lines. 1ofqHyPgr74Vx 0vUkETWFAIGNORECASEu8SJQ5C 1 vfC7IGNORECASEMUtRWYq3KGKJpR8koi7FhtzX _____________ OTMODZfX1gD9l38Tu9PEQZrshVzLIGNORECASEbI / Good Job! \ u7YtPPNnVLSzB8HCBvtOcIHey0X8WtIgnorEcase | You did a | QfX1XYVyUHpwUIgnorEcasepT fi6GkHvOkG LDb | case-insens | Vw4ePnDoZ4KxNs58pWlGMoFVcIgnorEcasepQj 6 | itive | fN4SOVBxl6IgnorEcaseeJ5Ldyb0y4PLVSL1ZCv7 \ search / mmNqW04FRacds3eYbIgnorEcaseRk5rFhFpKahDt ------------- ZgQZAYDnIE7Jk4PLhZ10gxIGNORECASEpxQqxB4t \ ^__^ 50FY1806ignOrecase6DzXRGwihWPeO3J gjHsDG \ (oo)\_______ QxAIpmflI jFcJignOrecaseQM06LNCSX lftJUX (__)\ )\/\ wignorecaseTt0lDGMb5KCFWEm4t8RmBNXtLvURX ||----w | w1EeylvQJWMFIgnorEcaseWavz4 ICR89dkvr6sf || || wayAmo30uEjxkMyJvisIGNORECASEkwshDQX DGB 45s7W ggfignOrecaseUYiHjY0F6BWSqqDfZ6c F zmqyIgnorEcaseqo5w9DIs0DGFlDayGlVaheoIlO
# To solve the exercise, we also have to count the number of output lines.
# This can be done with the "-c" option:
grep -i -c ignorecase data/exercise1_grep.txt
21
# solution: how to find the instructions for the next exercise
grep 21 data/exercise1_grep.txt
tidjh21yvuMNPDEma8t6PksdgTVkimf6F8LHegXf OllivZL3QFq8OiobDOQjdrPT1KeqT21 bRG WMRc eCmkBM21OATsb57fD9ao6czsMB1f7gtWvJCFAW3z ____________________ YCOQlk1yUmr8EjN3NBxEB0SSToh21Xfpm BiVHS7 / Congrats! Yes \ JCsq1gs3drLCHAerYroSp331AJMHr21m9Atm4UMR | the answer to the | z3nfFTpzSKGHfdDwtIadMjgiYx21iiat3S9VVT8R | case-insensitive | 0qBEpfp1dcTibKVwObda341CTH9zoYJpBFe821yy | and count question | KJIsvaofywLv6uz1216aZlUBQ3XBJd1jVC5bdHAE | is 21. / jy0FgakHM4Tq7ncjhUN21ggkNyZhNhJC4eyz ESN -------------------- xwwOmWdp5pJ8IsvtNMx9EnWOnjmuUEdt4o8d21zc \ ^__^ k azZdXgjRGFYTHuMIp0SFkwjp4vHRG1lnlmSj21 \ (oo)\_______ jYe19iH7NaYtPGDC7mXoy5G721s8EGrD8wFCZSlJ (__)\ )\/\ CXUYNxwnP8jr3NR5T9SCl5TQAwJI5ZjNCm zw21Y ||----w | l21FpLp HaLHc1MaoMXflHI4wr981PUNefC0cKDC || || fC1BsEyvpDm cCnceoQCj321v36bmPx5u9Ht6qxs VAAh4PTzYzWSbMxmtDE8XtwYqSu8KFq5021ycKLY WmHhzfH8XzJ4Dd3PvgMoIXAnoJJG3G9HlGUtD21d ============= yCrjC21uBDHKBR1P0XVXQp9XE6T7Nqa6C p8ZQ4H Next exercise zfa7If6rzhvuv O6HFHU21cbLnpW0Yipf3xSKJSS ============= FPgwt6n3mfTJtartXVwrMAtmn3ISF21yiK0U9NH4 PnV21lkRoTqqVP 9Hs4v4RlJLFdOx6LkhICM WW1 Searching in multiple files uaRh219wTTl0wCVin63cfrywW06LwQOb vx1k5Uu FTOHCTMDFlKj cNVu21DgKqN1EZxhU1iPyGRrko1 Grep can search the same pattern EfzALglVAh8cPso5WmyYi8v1QG0c21LUTKPqw66N in more than one file at the same time. rGWMTbnXJnehtyAY3vxTJWUdaUXH MxFnyA21fUN l0UcWWd0LG0GeFwNKlGEyj07pbUOPTee121t0MsN The folder data/multiplefiles/ contains hundreds of different files. Ow7gE6ZNvIGLP775npX6j5menzWz421Hg00qDP3w uWhIZ4kk6cI7d9503RXAniriZjemCOZ21J7BTCBt Can you identify the file containing the word "regex"? Wxd4621C JxW68aYMWbeCMY0eVMtTqF8iAfhqazV iekhxfE5LpZ21qUxwIjXpYtMchz489rzXtZ0 VOU
Searching multiple files¶
Grep is useful to search over multiple files in a single command.
The folder data/multiplefiles/ contains 50 randomly generated files. You can see their contents with head data/multiplefiles/* or with less.
One of these files contains the word "regex" in it. Are you able to find it?
# solution: you can use the "*" character to specify multiple files:
grep 'regex' data/multiplefiles/*
data/multiplefiles/file32.txt:5gsumFTKbKEJv9dD8W94FhoEQU8qf8RMUcregexR data/multiplefiles/file32.txt:YgDiqkA C1oregex9giqI66c3sOwfLirOsgPpSuq data/multiplefiles/file32.txt:IsXSnp 8U8pKR0LsVuKregexO5GFegOtV4GW4fNQ Good! You've found the data/multiplefiles/file32.txt:l 4px8KhPRmfEJgi5uTuVO1XahG3H1sYregex4wt file containing the word "regex" data/multiplefiles/file32.txt:yz8P5 HC6N5D XRHPncZjTAeMregexT9bQUoZdsh data/multiplefiles/file32.txt:eWUd18s0MVx5YYrEK KCKeF5hvOregexIiZbIGUX To continue, data/multiplefiles/file32.txt:MLXiKZJ8KyHMou9lYsz4ZjFYJSfB 14tregextpJ grep file32.txt data/exercise1_grep.txt data/multiplefiles/file32.txt:veFQUregexfnQxwQw6POJRNvvAeYwToX6ptvN39m data/multiplefiles/file32.txt:cHoNvregexiGjHkmptPVTjOzvWVGbrGoHoywV4Vy
Searching multiple patterns and the Unix piping system¶
How can we search that contain two or more patterns?
One solution is to use the Unix piping system, executing one grep command, and then another grep on the output.
This can be done using the pipe "|" symbol, like the following:
$: grep (first pattern) myfile.txt | grep (second pattern)
Press space or the down key for some examples.
The file data/genes/mgat_genes.gb is a genbank file. Notice how this format is well suited for grep searches:
head data/genes/mgat_genes.gb
LOCUS HUMUDPCNA 4705 bp DNA linear PRI 19-SEP-1995
DEFINITION Human alpha-1,3-mannosyl-glycoprotein beta-1,
2-N-acetylglucosaminyltransferase (MGAT) gene, complete cds.
ACCESSION M61829
VERSION M61829.1 GI:340075
KEYWORDS alpha-1,3-mannosyl-glycoprotein beta-1,2-N-acetylglucosaminyltrae.
SOURCE Homo sapiens (human)
ORGANISM Homo sapiens
Eukaryota; Metazoa; Chordata; Craniata; Vertebrata; Euteleostomi;
Mammalia; Eutheria; Euarchontoglires; Primates; Haplorrhini;
Let's say we want to search all the lines where "ORGANISM" is "Homo sapiens".
We can do it with two grep commands:
grep ORGANISM data/genes/mgat_genes.gb | grep 'Homo sapiens'
Notice that searching for "Homo sapiens" alone would not be enough, as there are other lines where the word "Homo sapiens" is present.
grep ORGANISM data/genes/mgat_genes.gb | grep 'Homo sapiens'
ORGANISM Homo sapiens ORGANISM Homo sapiens ORGANISM Homo sapiens ORGANISM Homo sapiens ORGANISM Homo sapiens ORGANISM Homo sapiens ORGANISM Homo sapiens ORGANISM Homo sapiens
The file contains sequences from two other organisms apart from Homo sapiens. Can you guess which one to search for the next exercise?
# Solution: grep for "bos taurus":
grep ORGANISM data/genes/mgat_genes.gb | grep taurus
ORGANISM Bos taurus ORGANISM Bos taurus ORGANISM Bos taurus ORGANISM Bos taurus _______________ ORGANISM Bos taurus < Well guessed! > ORGANISM Bos taurus --------------- ORGANISM Bos taurus \ ^__^ ORGANISM Bos taurus \ (oo)\_______ ORGANISM Bos taurus (__)\ )\/\ ORGANISM Bos taurus ||----w | ORGANISM Bos taurus || || ORGANISM Bos taurus ORGANISM Bos taurus ORGANISM Bos taurus ORGANISM Bos taurus ORGANISM Bos taurus ORGANISM Bos taurus =============== ORGANISM Bos taurus Next Exercise ORGANISM Bos taurus =============== ORGANISM Bos taurus ORGANISM Bos taurus To continue, grep ORGANISM Bos taurus "regex" in ORGANISM Bos taurus data/exercise1_grep.txt ORGANISM Bos taurus
Regular Expressions¶
Regular expressions allow to search for more complex patterns.
Here are some simple regular expression examples:
| regex | description |
|---|---|
| . | matches any character |
| [A-Za-z] | matches any of the characters within parenthesis |
| .* | matches any character, any number of times |
Regular Expression exercise¶
Let's have a look at the file data/genes/sequences.fasta:
head data/genes/sequences.fasta
>seq000 sequence description NANTCNNNGNATNNCNTACNTGNTCGNCCG >seq001 sequence description TGTCAATTNTCNGCGTCNNACNNACTCGCN >seq002 sequence description TGGGCGNTCATGNANAATGTTACGCTCNGG >seq003 sequence description GCCTTTNGGNNCTCACTANGCANGTTTGAN >seq004 sequence description CATNANNAAAccTTTAGGCACTCNACACNG
Can you use grep to identify all the sequences containing three As, followed by any two characters, followed by three Ts?
grep 'AAA..TTT' data/genes/sequences.fasta
CATNANNAAAccTTTAGGCACTCNACACNG AGGCCGCNGNGGTAAAActTTTACNAAGAC GTGTNNGTCAAGCNCGNCGTTNAAAGGTTT ATNCGNAGNNCANTNGACAAAccTTTTTGT NTACNTAAAAgtTTTCCACTNTTTANTCAA CNNGAGCGAAActTTTNGCAAGTCTGNNCN CATGGGCAAAgtTTTATANATGTNAANCNT GGTGGGNNCCCAGNCGNCAAAgtTTTNNCT GAAAAgtTTTTCNAACTTTNNAATAANNCN GAACAAGCNGCCCTTGGCCAAAgtTTTGNC NNTCGTNGNNNAAAAaaTTTTAAGAGCACC NNNGNGTNGAAActTTTTTGAACGANNAAT CCAAAaaTTTNTAGNAGCCTGTAGAGCCGC
Bonus: if we use the -B 1 grep option, we can retrieve the names of these sequences:
grep -B1 'AAA..TTT' data/genes/sequences.fasta
>seq004 sequence description CATNANNAAAccTTTAGGCACTCNACACNG -- >seq012 sequence description ________________ AGGCCGCNGNGGTAAAActTTTACNAAGAC -- >seq015 sequence description GTGTNNGTCAAGCNCGNCGTTNAAAGGTTT -- >seq022 sequence description / Congrats! This \ ATNCGNAGNNCANTNGACAAAccTTTTTGT >seq023 sequence description | was the last | NTACNTAAAAgtTTTCCACTNTTTANTCAA -- >seq029 sequence description \ grep exercise / CNNGAGCGAAActTTTNGCAAGTCTGNNCN >seq030 sequence description ---------------- CATGGGCAAAgtTTTATANATGTNAANCNT >seq031 sequence description \ ^__^ GGTGGGNNCCCAGNCGNCAAAgtTTTNNCT -- >seq033 sequence description \ (oo)\_______ GAAAAgtTTTTCNAACTTTNNAATAANNCN >seq034 sequence description (__)\ )\/\ GAACAAGCNGCCCTTGGCCAAAgtTTTGNC -- >seq038 sequence description ||----w | NNTCGTNGNNNAAAAaaTTTTAAGAGCACC >seq039 sequence description || || NNNGNGTNGAAActTTTTTGAACGANNAAT -- >seq041 sequence description CCAAAaaTTTNTAGNAGCCTGTAGAGCCGC
# Bonus: pipe an additional grep '>' to see a cow:
grep -B1 'AAA..TTT' data/genes/sequences.fasta | grep '>'
>seq004 sequence description >seq012 sequence description ________________ >seq015 sequence description >seq022 sequence description / Congrats! This \ >seq023 sequence description | was the last | >seq029 sequence description \ grep exercise / >seq030 sequence description ---------------- >seq031 sequence description \ ^__^ >seq033 sequence description \ (oo)\_______ >seq034 sequence description (__)\ )\/\ >seq038 sequence description ||----w | >seq039 sequence description || || >seq041 sequence description
Working with tabular files: Awk¶
The awk command allows to search and manipulate tabular files from the command line.
Imagine it as the equivalent of Excel/Calc for the command line. It allows to do search on specific columns of a file, to do numerical operations, or to change the order of the columns.
The advantage of a command-line tool over graphical software is that the memory footprint is much lower. So you can access and modify large files in a fraction of the time that it would take with Excel.
Example of tabular file: the GFF3 format¶
The file data/genes/chr8.gff contains an example of file in the GFF3 format:
head data/genes/chr8.gff
##gff-version 3 ##source-version refgene 1.28.10 ##date 2016-09-08 ##genome-build . hg19 chr8 refgene gene 18248755 18258723 . + . gene_id=10;symbol=NAT2;;ID=10 chr8 refgene gene 100549014 100549089 . - . gene_id=100126309;symbol=MIR875;;ID=100126309 chr8 refgene gene 144895127 144895212 . - . gene_id=100126338;symbol=MIR937;;ID=100126338 chr8 refgene gene 145619364 145619445 . - . gene_id=100126351;symbol=MIR939;;ID=100126351 chr8 refgene gene 91970706 91997485 . - . gene_id=100127983;symbol=C8orf88;;ID=100127983 chr8 refgene gene 74332309 74353753 . + . gene_id=100128126;symbol=STAU2-AS1;;ID=100128126
As you can see it is a tab-separated file, which we could easily read in Excel or Calc.
The format specifications are defined here, but in short:
- the first, fourth and fifth columns contain the chromosome name and coordinates
- the second column describes the tool or resource that generated the annotation
- the third column describe the type of feature (e.g. gene, transcript, exon, TF binding site, Histone Acetylation mark, etc...
- the ninth column contains several fields, separated by a semicolon
Basic AWK syntax: filters¶
The basic AWK syntax is the following:
awk 'filters {print statements}' filename
Awk is quite smart at recognizing the field separator, and by default assumes they are separated by tabs.
Each column of the file can be referred to with the dollar sign followed by the number of column.
For example $2 refers to the second column, and so on.
The following code filters all the lines belonging to chromosome 8, between the coordinates 100000 and 200000:
awk '$1=="chr8" && $4>100000 && $5<200000 ' data/genes/chr8.gff
chr8 refgene gene 182200 197339 . + . gene_id=169270;symbol=ZNF596;;ID=169270 chr8 refgene gene 116086 117024 . - . gene_id=441308;symbol=OR4F21;;ID=441308 chr8 refgene gene 158345 182318 . - . gene_id=644128;symbol=RPL23AP53;;ID=644128
Exercise¶
Can you print all the lines between 5000000 and 10000000 ?
awk '$4 > 5000000 && $5 < 10000000 ' data/genes/chr8.gff
chr8 refgene gene 7143733 7212876 . - . gene_id=100128890;symbol=FAM66B;ID=100128890 chr8 refgene gene 7215498 7220490 . - . gene_id=100131980;symbol=ZNF705G;ID=100131980 chr8 refgene gene 7812535 7866277 . + . gene_id=100132103;symbol=FAM66E;ID=100132103 chr8 refgene gene 7783859 7809935 . + . _________ chr8 refgene gene 6261077 6264069 . - . / Cows in \ chr8 refgene gene 7272385 7274354 . - . | the | chr8 refgene gene 7946463 7946611 . - . \ Genome! / chr8 refgene gene 6602685 6602765 . + . --------- chr8 refgene gene 8905955 8906028 . + . \ ^__^ chr8 refgene gene 6602689 6602761 . - . \ (oo)\_______ chr8 refgene gene 6693076 6699975 . + . (__)\ )\/\ chr8 refgene gene 8559666 8561617 . + . ||----w | chr8 refgene gene 9182561 9192590 . + . || | chr8 refgene gene 8175258 8239257 . - . gene_id=157285;symbol=SGK223;ID=157285 chr8 refgene gene 9757574 9760839 . - . gene_id=157627;symbol=LINC00599;ID=157627 chr8 refgene gene 6835171 6856724 . - . gene_id=1667;symbol=DEFA1;ID=1667 chr8 refgene gene 6793345 6795786 . - . gene_id=1669;symbol=DEFA4;ID=1669 chr8 refgene gene 6912829 6914259 . - . gene_id=1670;symbol=DEFA5;ID=1670 chr8 refgene gene 6782216 6783598 . - . gene_id=1671;symbol=DEFA6;ID=1671 chr8 refgene gene 6728097 6735529 . - . gene_id=1672;symbol=DEFB1;ID=1672 chr8 refgene gene 7752199 7754237 . + . gene_id=1673;symbol=DEFB4A;ID=1673 chr8 refgene gene 6844700 6866346 . - . gene_id=170949;symbol=DEFT1P;ID=170949 chr8 refgene gene 7353368 7366833 . + . gene_id=245910;symbol=DEFB107A;ID=245910 chr8 refgene gene 6357175 6420784 . - . gene_id=285;symbol=ANGPT2;ID=285 chr8 refgene gene 8086092 8102387 . + . gene_id=286042;symbol=FAM86B3P;ID=286042 chr8 refgene gene 6666041 6693166 . - . gene_id=389610;symbol=XKR5;ID=389610 chr8 refgene gene 7829183 7830775 . - . gene_id=392188;symbol=USP17L8;ID=392188 chr8 refgene gene 7189909 7191501 . + . gene_id=401447;symbol=USP17L1;ID=401447 chr8 refgene gene 9760898 9760982 . - . gene_id=406907;symbol=MIR124-1;ID=406907 chr8 refgene gene 7413660 7431920 . - . gene_id=441317;symbol=FAM90A7P;ID=441317 chr8 refgene gene 7627106 7628835 . + . gene_id=441328;symbol=FAM90A10P;ID=441328 chr8 refgene gene 6808248 6809121 . - . gene_id=449491;symbol=DEFA8P;ID=449491 chr8 refgene gene 6816811 6817683 . - . gene_id=449492;symbol=DEFA9P;ID=449492 chr8 refgene gene 6825663 6826635 . - . gene_id=449493;symbol=DEFA10P;ID=449493 chr8 refgene gene 7669242 7673238 . - . gene_id=503614;symbol=DEFB107B;ID=503614 chr8 refgene gene 6565878 6619021 . + . gene_id=55326;symbol=AGPAT5;ID=55326 chr8 refgene gene 7194637 7196229 . + . gene_id=645402;symbol=USP17L4;ID=645402 chr8 refgene gene 7833915 7835507 . - . gene_id=645836;symbol=USP17L3;ID=645836 chr8 refgene gene 7705402 7721319 . + . gene_id=653423;symbol=SPAG11A;ID=653423 chr8 refgene gene 9599182 9599278 . + . gene_id=693182;symbol=MIR597;ID=693182 chr8 refgene gene 6886123 6887011 . - . gene_id=724068;symbol=DEFA11P;ID=724068 chr8 refgene gene 6873391 6875823 . - . gene_id=728358;symbol=DEFA1B;ID=728358 chr8 refgene gene 6264113 6501140 . + . gene_id=79648;symbol=MCPH1;ID=79648 chr8 refgene gene 8993764 9009152 . - . gene_id=79660;symbol=PPP1R3B;ID=79660 chr8 refgene gene 9413445 9639856 . + . gene_id=8658;symbol=TNKS;ID=8658 chr8 refgene gene 8860314 8890849 . + . gene_id=90459;symbol=ERI1;ID=90459 chr8 refgene gene 8641999 8751131 . - . gene_id=9258;symbol=MFHAS1;ID=9258
Awk: printing columns and doing operations¶
Awk also allows to print only specific columns, and do algebraic operations on them.
Remember that each column can be referred as $1, $2, $3, etc...
For example the following code prints the first column, and the sum of the fourth and third. We can pipe the output to head or less, to make it easier to visualize:
awk '{print $1, $5-$4}' data/genes/chr8.gff | head
##gff-version 0 ##source-version 0 ##date 0 ##genome-build 0 chr8 9968 chr8 75 chr8 85 chr8 81 chr8 26779 chr8 21444 awk: write failure (Broken pipe) awk: close failed on file /dev/stdout (Broken pipe)
Notice how this also prints the headers of the file. We can exclude these by adding a grep condition:
awk '{print $1, $5-$4, $9}' data/genes/chr8.gff | grep -v '^#' | head
chr8 9968 gene_id=10;symbol=NAT2;;ID=10 chr8 75 gene_id=100126309;symbol=MIR875;;ID=100126309 chr8 85 gene_id=100126338;symbol=MIR937;;ID=100126338 chr8 81 gene_id=100126351;symbol=MIR939;;ID=100126351 chr8 26779 gene_id=100127983;symbol=C8orf88;;ID=100127983 chr8 21444 gene_id=100128126;symbol=STAU2-AS1;;ID=100128126 chr8 12197 gene_id=100128338;symbol=FAM83H-AS1;;ID=100128338 chr8 1835 gene_id=100128627;symbol=CDC42P3;;ID=100128627 chr8 3282 gene_id=100128750;symbol=RBPMS-AS1;;ID=100128750 chr8 69143 gene_id=100128890;symbol=FAM66B;ID=100128890
Exercise (difficult)¶
Starting from the previous command, can you extract the gene symbol into a separate column?
Hints: pipe an additional awk statement after the first. Use the -F option to specify a different field separator.
awk '{print $1, $5-$4, $9}' data/genes/chr8.gff | grep -v '^#' | awk -F';' '{print $1, $2}' | head
chr8 9968 gene_id=10 symbol=NAT2 chr8 75 gene_id=100126309 symbol=MIR875 chr8 85 gene_id=100126338 symbol=MIR937 chr8 81 gene_id=100126351 symbol=MIR939 chr8 26779 gene_id=100127983 symbol=C8orf88 chr8 21444 gene_id=100128126 symbol=STAU2-AS1 chr8 12197 gene_id=100128338 symbol=FAM83H-AS1 chr8 1835 gene_id=100128627 symbol=CDC42P3 chr8 3282 gene_id=100128750 symbol=RBPMS-AS1 chr8 69143 gene_id=100128890 symbol=FAM66B awk: write failure (Broken pipe) awk: close failed on file /dev/stdout (Broken pipe) grep: write error
AWK: searching by regular expressions¶
Awk can also be used to search by regular expression.
For example, the following code will print all the lines in which the symbol starts with "MIR":
awk '$9 ~ /symbol=MIR/ {print $0}' data/genes/chr8.gff
chr8 refgene gene 100549014 100549089 . - . gene_id=100126309;symbol=MIR875;;ID=100126309 chr8 refgene gene 144895127 144895212 . - . gene_id=100126338;symbol=MIR937;;ID=100126338 chr8 refgene gene 145619364 145619445 . - . gene_id=100126351;symbol=MIR939;;ID=100126351 chr8 refgene gene 65285775 65295842 . + . gene_id=100130155;symbol=MIR124-2HG;;ID=100130155 chr8 refgene gene 128972879 128972941 . + . gene_id=100302161;symbol=MIR1205;;ID=100302161 chr8 refgene gene 10682883 10682953 . - . gene_id=100302166;symbol=MIR1322;;ID=100302166 chr8 refgene gene 129021144 129021202 . + . gene_id=100302170;symbol=MIR1206;;ID=100302170 chr8 refgene gene 129061398 129061484 . + . gene_id=100302175;symbol=MIR1207;;ID=100302175 chr8 refgene gene 128808208 128808274 . + . gene_id=100302185;symbol=MIR1204;;ID=100302185 chr8 refgene gene 145625476 145625559 . - . gene_id=100302196;symbol=MIR1234;;ID=100302196 chr8 refgene gene 113655722 113655812 . + . gene_id=100302225;symbol=MIR2053;;ID=100302225 chr8 refgene gene 27743556 27743633 . - . gene_id=100422828;symbol=MIR4287;;ID=100422828 chr8 refgene gene 29814788 29814864 . - . gene_id=100422876;symbol=MIR3148;;ID=100422876 chr8 refgene gene 28362633 28362699 . - . gene_id=100422903;symbol=MIR4288;;ID=100422903 chr8 refgene gene 96085142 96085221 . + . gene_id=100422964;symbol=MIR3150A;;ID=100422964 chr8 refgene gene 104166842 104166917 . + . gene_id=100422992;symbol=MIR3151;;ID=100422992 chr8 refgene gene 12584746 12584808 . + . gene_id=100500838;symbol=MIR3926-2;;ID=100500838 chr8 refgene gene 27559194 27559276 . + . gene_id=100500858;symbol=MIR3622A;;ID=100500858 chr8 refgene gene 12584741 12584813 . - . gene_id=100500870;symbol=MIR3926-1;;ID=100500870 chr8 refgene gene 27559190 27559284 . - . gene_id=100500871;symbol=MIR3622B;;ID=100500871 chr8 refgene gene 96085139 96085224 . - . gene_id=100500907;symbol=MIR3150B;;ID=100500907 chr8 refgene gene 117886967 117887039 . - . gene_id=100500914;symbol=MIR3610;;ID=100500914 chr8 refgene gene 42751340 42751418 . - . gene_id=100616115;symbol=MIR4469;;ID=100616115 chr8 refgene gene 94928250 94928347 . - . gene_id=100616169;symbol=MIR378D2;;ID=100616169 chr8 refgene gene 29920258 30108213 . - . gene_id=100616190;symbol=MIR548O2;;ID=100616190 chr8 refgene gene 92217713 92217786 . + . gene_id=100616245;symbol=MIR4661;;ID=100616245 chr8 refgene gene 124228028 124228103 . - . gene_id=100616260;symbol=MIR4663;;ID=100616260 chr8 refgene gene 143257700 143257779 . + . gene_id=100616268;symbol=MIR4472-1;;ID=100616268 chr8 refgene gene 144815253 144815323 . - . gene_id=100616318;symbol=MIR4664;;ID=100616318 chr8 refgene gene 101394991 101395073 . + . gene_id=100616451;symbol=MIR4471;;ID=100616451 chr8 refgene gene 62627347 62627418 . + . gene_id=100616484;symbol=MIR4470;;ID=100616484 chr8 refgene gene 103137660 103137743 . + . gene_id=100847001;symbol=MIR5680;;ID=100847001 chr8 refgene gene 131020580 131020699 . - . gene_id=100847051;symbol=MIR5194;;ID=100847051 chr8 refgene gene 81153624 81153708 . + . gene_id=100847056;symbol=MIR5708;;ID=100847056 chr8 refgene gene 75460778 75460852 . + . gene_id=100847058;symbol=MIR5681A;;ID=100847058 chr8 refgene gene 75460785 75460844 . - . gene_id=100847091;symbol=MIR5681B;;ID=100847091 chr8 refgene gene 9760898 9760982 . - . gene_id=406907;symbol=MIR124-1;ID=406907 chr8 refgene gene 65291706 65291814 . + . gene_id=406908;symbol=MIR124-2;;ID=406908 chr8 refgene gene 135812763 135812850 . - . gene_id=407030;symbol=MIR30B;;ID=407030 chr8 refgene gene 135817119 135817188 . - . gene_id=407033;symbol=MIR30D;;ID=407033 chr8 refgene gene 22102475 22102556 . - . gene_id=407037;symbol=MIR320A;;ID=407037 chr8 refgene gene 75512101 75670587 . + . gene_id=441355;symbol=MIR2052HG;;ID=441355 chr8 refgene gene 14710947 14711019 . - . gene_id=494332;symbol=MIR383;;ID=494332 chr8 refgene gene 41517959 41518026 . - . gene_id=619554;symbol=MIR486-1;;ID=619554 chr8 refgene gene 1765397 1765473 . + . gene_id=693181;symbol=MIR596;;ID=693181 chr8 refgene gene 9599182 9599278 . + . gene_id=693182;symbol=MIR597;ID=693182 chr8 refgene gene 10892716 10892812 . - . gene_id=693183;symbol=MIR598;;ID=693183 chr8 refgene gene 100548864 100548958 . - . gene_id=693184;symbol=MIR599;;ID=693184 chr8 refgene gene 145019359 145019447 . - . gene_id=724031;symbol=MIR661;;ID=724031
Last exercise!¶
Calculate the lenght of the gene POU5F1B.
Find the Gene whose gene_id is equal to that number.
awk '$9 ~ /POU5F1B/ {print $5-$4}' data/genes/chr8.gff
1584
awk '$9 ~ /gene_id=1584/ {print $0}' data/genes/chr8.gff
chr8 refgene Good_Job! 143953773 143961236 . - . gene_id=1584;symbol=CYP11B1;;ID=1584
Bonus: Makefiles¶
Let's have a look at the file called Makefile in the exercise directory:
head Makefile
test_exercises: start help ignorecase multiplefiles generate_exercises: generate_grep generate_awk testrule: echo this is a Makefile rule echo You can associate it to as many commands you want notebook: jupyter nbconvert --to notebook --execute PEB\ Bash\ Workshop.ipynb
Press space or the down key to continue
Defining pipelines with Makefiles¶
Makefiles are a basic way to define pipelines of shell commands.
Nowadays there are more sophisticated tools available, but most of these are based on Makefiles.
A Makefile is a collection of "rules".
Each of these rules follows this basic syntax is:
target: prerequisites
commands to execute
As you can see in the Makefile included, most of the rules allow to regenerate the exercise files, or to execute some commands without having to type them everytime.
For example, the rule "testrule" is associated to two echo commands.
How to run Makefile rules¶
To execute a rule in the Makefile, simply type:
make [name of the rule]
For example:
make testrule
echo this is a Makefile rule this is a Makefile rule echo You can associate it to as many commands you want You can associate it to as many commands you want
The program "make" will automatically detect any file named "Makefile" in the current directory, and execute any rule with the specific name.
Rules can also be nested together. For example the two rules "test_exercises" and "generate_exercises" at the beginning of the file are a way to call several other rules together.
The last slide¶
This is the last slide of the workshop. To finish, try to execute the rule "cow" in the Makefile.
make cow
_____________
/ I hope you \
| have |
| enjoyed the |
| workshop |
\ :-) /
-------------
\ ^__^
\ (oo)\_______
(__)\ )\/\
||----w |
|| ||
___________
( Now let's )
( go to the )
( beach )
-----------
o ^__^
o (oo)\_______
(__)\ )\/\
||----w |
|| ||