#!/usr/bin/env python
# coding: utf-8
#
#
# # Demo: Probabilistic neural network training for denoising of synthetic 2D data
#
# This notebook demonstrates training a probabilistic CARE model for a 2D denoising task, using provided synthetic training data.
# Note that training a neural network for actual use should be done on more (representative) data and with more training time.
#
# More documentation is available at http://csbdeep.bioimagecomputing.com/doc/.
# In[1]:
from __future__ import print_function, unicode_literals, absolute_import, division
import numpy as np
import matplotlib.pyplot as plt
get_ipython().run_line_magic('matplotlib', 'inline')
get_ipython().run_line_magic('config', "InlineBackend.figure_format = 'retina'")
from tifffile import imread
from csbdeep.utils import download_and_extract_zip_file, axes_dict, plot_some, plot_history
from csbdeep.utils.tf import limit_gpu_memory
from csbdeep.io import load_training_data
from csbdeep.models import Config, CARE
# The TensorFlow backend uses all available GPU memory by default, hence it can be useful to limit it:
# In[2]:
# limit_gpu_memory(fraction=1/2)
#
#
# # Training data
#
# Download and read provided training data, use 10% as validation data.
# In[3]:
download_and_extract_zip_file (
url = 'http://csbdeep.bioimagecomputing.com/example_data/synthetic_disks.zip',
targetdir = 'data',
)
# In[4]:
(X,Y), (X_val,Y_val), axes = load_training_data('data/synthetic_disks/data.npz', validation_split=0.1, verbose=True)
c = axes_dict(axes)['C']
n_channel_in, n_channel_out = X.shape[c], Y.shape[c]
# In[5]:
plt.figure(figsize=(12,5))
plot_some(X_val[:5],Y_val[:5])
plt.suptitle('5 example validation patches (top row: source, bottom row: target)');
#
#
# # CARE model
#
# Before we construct the actual CARE model, we have to define its configuration via a `Config` object, which includes
# * parameters of the underlying neural network,
# * the learning rate,
# * the number of parameter updates per epoch,
# * the loss function, and
# * whether the model is probabilistic or not.
#
# The defaults should be sensible in many cases, so a change should only be necessary if the training process fails.
#
# For a probabilistic model, we have to explicitly set `probabilistic=True`.
#
# ---
#
# Important: Note that for this notebook we use a very small number of update steps per epoch for immediate feedback, whereas this number should be increased considerably (e.g. `train_steps_per_epoch=400`) to obtain a well-trained model.
# In[6]:
config = Config(axes, n_channel_in, n_channel_out, probabilistic=True, train_steps_per_epoch=30)
print(config)
vars(config)
# We now create a CARE model with the chosen configuration:
# In[7]:
model = CARE(config, 'my_model', basedir='models')
#
#
# # Training
#
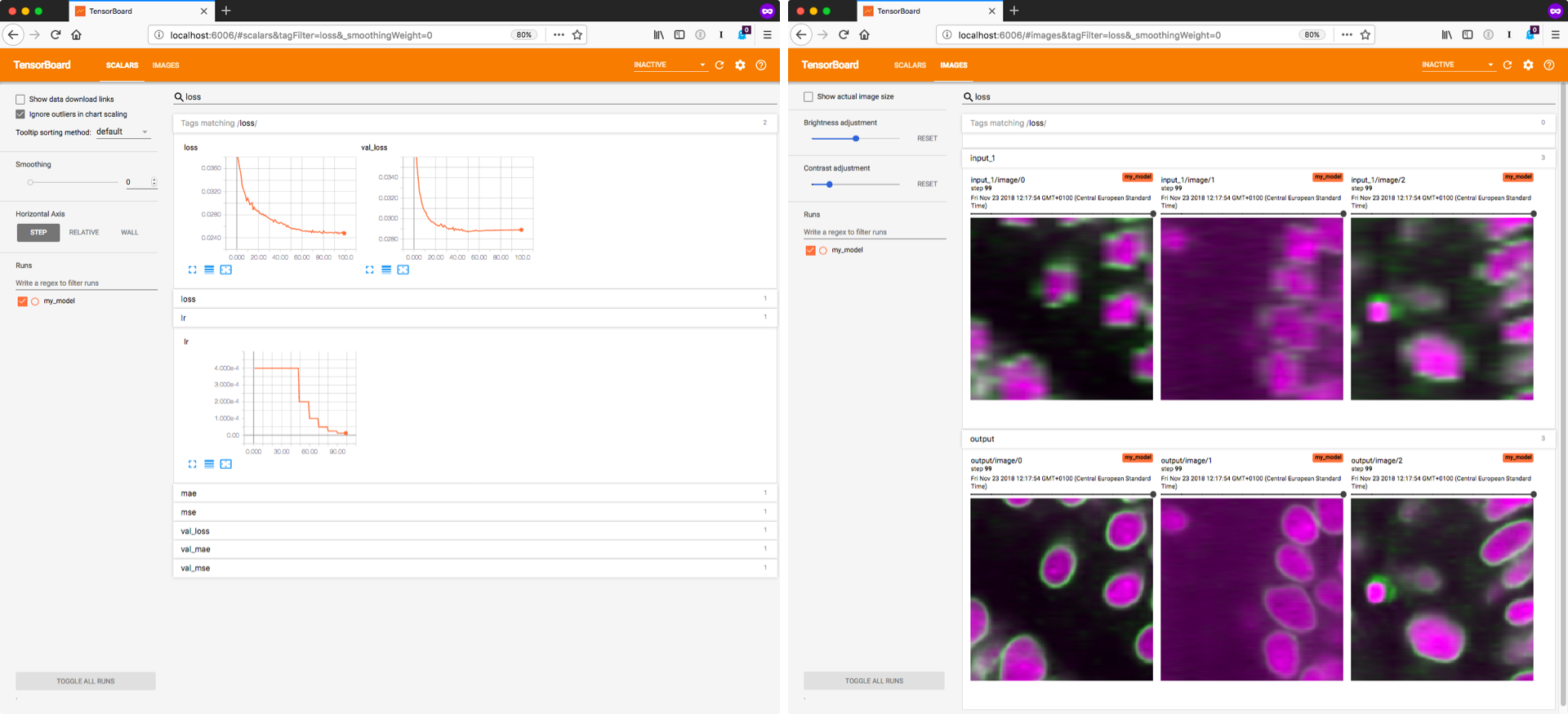
# Training the model will likely take some time. We recommend to monitor the progress with [TensorBoard](https://www.tensorflow.org/programmers_guide/summaries_and_tensorboard) (example below), which allows you to inspect the losses during training.
# Furthermore, you can look at the predictions for some of the validation images, which can be helpful to recognize problems early on.
#
# You can start TensorBoard from the current working directory with `tensorboard --logdir=.`
# Then connect to [http://localhost:6006/](http://localhost:6006/) with your browser.
#
# 
# In[8]:
history = model.train(X,Y, validation_data=(X_val,Y_val))
# Plot final training history (available in TensorBoard during training):
# In[9]:
print(sorted(list(history.history.keys())))
plt.figure(figsize=(16,5))
plot_history(history,['loss','val_loss'],['mse','val_mse','mae','val_mae']);
#
#
# # Evaluation
#
# Example results for validation images.
# In[10]:
plt.figure(figsize=(12,10))
_P = model.keras_model.predict(X_val[:5])
_P_mean = _P[...,:(_P.shape[-1]//2)]
_P_scale = _P[...,(_P.shape[-1]//2):]
plot_some(X_val[:5],Y_val[:5],_P_mean,_P_scale,pmax=99.5)
plt.suptitle('5 example validation patches\n'
'first row: input (source), '
'second row: target (ground truth), '
'third row: predicted Laplace mean, '
'forth row: predicted Laplace scale');
#
#
# # Export model to be used with CSBDeep **Fiji** plugins and **KNIME** workflows
#
# See https://github.com/CSBDeep/CSBDeep_website/wiki/Your-Model-in-Fiji for details.
# In[11]:
model.export_TF()