#!/usr/bin/env python
# coding: utf-8
# ### From the Titanic Dataset
# 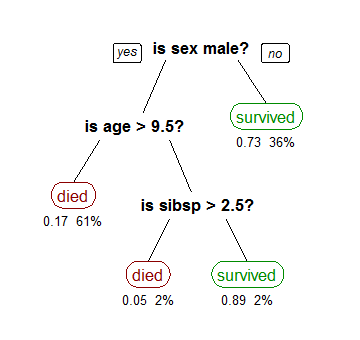
# By Stephen Milborrow (Own work) [CC BY-SA 3.0 via Wikimedia Commons]
# In[1]:
import mglearn # credits to Muller and Guido (https://www.amazon.com/dp/1449369413/)
import matplotlib.pyplot as plt
import numpy as np
get_ipython().run_line_magic('matplotlib', 'inline')
mglearn.plots.plot_tree_not_monotone()
# In[2]:
from sklearn.datasets import load_breast_cancer
from sklearn.tree import DecisionTreeClassifier
from sklearn.model_selection import train_test_split
cancer = load_breast_cancer()
X_train, X_test, y_train, y_test = train_test_split(cancer.data, cancer.target, stratify=cancer.target, random_state=42)
tree = DecisionTreeClassifier(random_state=0)
tree.fit(X_train, y_train)
print('Accuracy on the training subset: {:.3f}'.format(tree.score(X_train, y_train)))
print('Accuracy on the test subset: {:.3f}'.format(tree.score(X_test, y_test)))
# In[3]:
tree = DecisionTreeClassifier(max_depth=4, random_state=0)
tree.fit(X_train, y_train)
print('Accuracy on the training subset: {:.3f}'.format(tree.score(X_train, y_train)))
print('Accuracy on the test subset: {:.3f}'.format(tree.score(X_test, y_test)))
# In[4]:
import graphviz
from sklearn.tree import export_graphviz
export_graphviz(tree, out_file='cancertree.dot', class_names=['malignant', 'benign'], feature_names=cancer.feature_names,
impurity=False, filled=True)
# 
# In[5]:
print('Feature importances: {}'.format(tree.feature_importances_))
type(tree.feature_importances_)
# In[6]:
print(cancer.feature_names)
# In[7]:
n_features = cancer.data.shape[1]
plt.barh(range(n_features), tree.feature_importances_, align='center')
plt.yticks(np.arange(n_features), cancer.feature_names)
plt.xlabel('Feature Importance')
plt.ylabel('Feature')
plt.show()
# ### Advantages of Decision Trees
#
# - easy to view and understand
# - no need to pre-process, normalize, scale, and/or standardize features
#
# ### Paramaters to work with
#
# - max_depth
# - min_samples_leaf, max_samples_leaf
# - max_leaf_nodes
# - etc.
#
# ### Main Disadvantages
#
# - tendency to overfit
# - poor generalization
#
# #### Possible work-around: ensembles of decision trees
# In[ ]: