#!/usr/bin/env python
# coding: utf-8
# # Intersect bed on features
# Based on notebook 04 - but separating out to see if there is difference between hypo and hyper methylated.
#  # In[2]:
get_ipython().system('date')
# In[20]:
get_ipython().run_line_magic('pylab', 'inline')
import scipy.stats as stats
# Feature (from nb -03)
#
# **tldr** 4 "new" tracks
#
# In[2]:
get_ipython().system('date')
# In[20]:
get_ipython().run_line_magic('pylab', 'inline')
import scipy.stats as stats
# Feature (from nb -03)
#
# **tldr** 4 "new" tracks
#  # ```
# /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf
# /Users/sr320/data-genomic/tentacle/rebuilt.gtf
# /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff
# /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff
# ```
# # DEGs
# `-wb Write the original entry in B for each overlap. Useful for knowing what A overlaps. Restricted by -f and -r.`
# tldr
#
#
# ```
# /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf
# /Users/sr320/data-genomic/tentacle/rebuilt.gtf
# /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff
# /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff
# ```
# # DEGs
# `-wb Write the original entry in B for each overlap. Useful for knowing what A overlaps. Restricted by -f and -r.`
# tldr
#
#  # ## Separating HYPO and HYPER
# In[1]:
get_ipython().system('head ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph')
# In[1]:
get_ipython().system('fgrep -c "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph')
# In[3]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph | head')
# In[4]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph')
# In[6]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph')
# In[7]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph')
# In[ ]:
# In[ ]:
# In[ ]:
# ### HYPO
# In[5]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[9]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[10]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[10]:
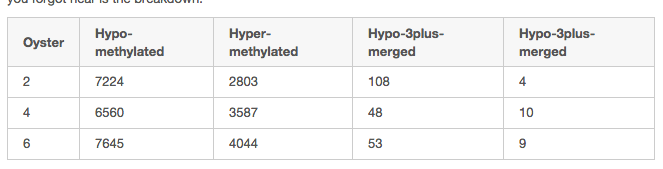
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 11 | sort | uniq -c')
# In[21]:
# Enter the data comparing Oyster 2 then Probes
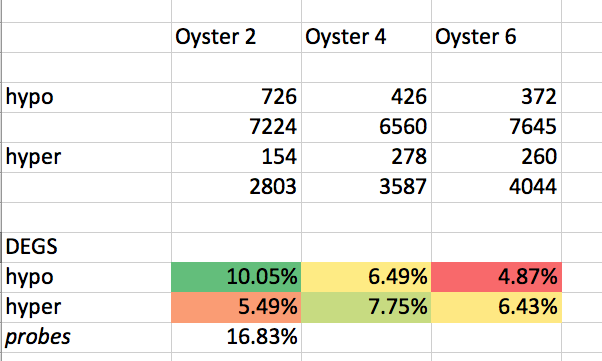
obs = array([[726, 7224], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[22]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[426, 6560], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[23]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[372, 7645], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# ## HYPER
# In[13]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
# In[12]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
# In[14]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
# In[15]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[16]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[17]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[18]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 11 | sort | uniq -c')
# In[24]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[154, 2803], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[25]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[278, 3587], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[26]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[260, 4044], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[ ]:
# In[ ]:
# In[ ]:
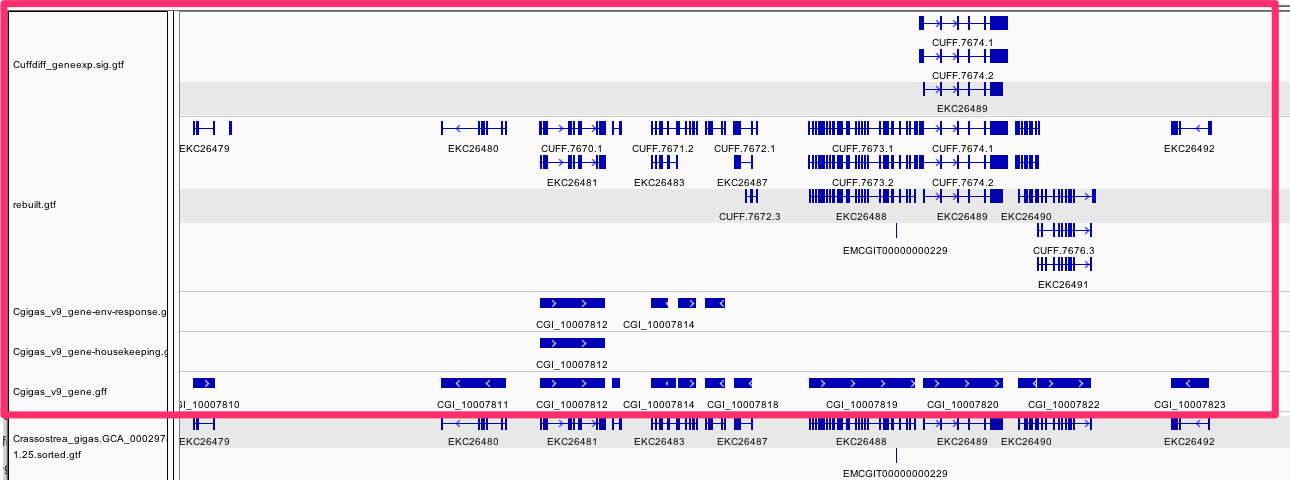
# # Rebuilt (new gtf based on RNAseq data)
# In[15]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[16]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[17]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[18]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 11 | sort | uniq -c')
# In[39]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[8768, 10028], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[40]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[7694, 10148], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[41]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[6160, 11690], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
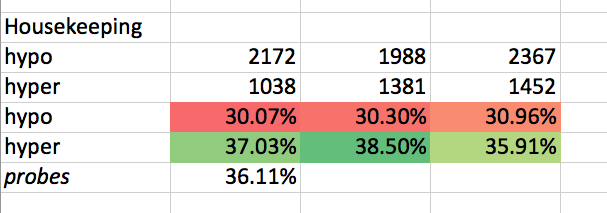
# # Housekeeping Genes
# Separating out hypo and hyper
#
# ## Separating HYPO and HYPER
# In[1]:
get_ipython().system('head ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph')
# In[1]:
get_ipython().system('fgrep -c "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph')
# In[3]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph | head')
# In[4]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph')
# In[6]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph')
# In[7]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph')
# In[ ]:
# In[ ]:
# In[ ]:
# ### HYPO
# In[5]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[9]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[10]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[10]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 11 | sort | uniq -c')
# In[21]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[726, 7224], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[22]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[426, 6560], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[23]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[372, 7645], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# ## HYPER
# In[13]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
# In[12]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
# In[14]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
# In[15]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[16]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[17]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[18]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 11 | sort | uniq -c')
# In[24]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[154, 2803], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[25]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[278, 3587], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[26]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[260, 4044], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[ ]:
# In[ ]:
# In[ ]:
# # Rebuilt (new gtf based on RNAseq data)
# In[15]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[16]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[17]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[18]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 11 | sort | uniq -c')
# In[39]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[8768, 10028], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[40]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[7694, 10148], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[41]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[6160, 11690], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# # Housekeeping Genes
# Separating out hypo and hyper
#  # In[29]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[30]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[31]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[26]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 11 | sort | uniq -c')
# In[42]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[3210, 10028], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[43]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[3369, 10148], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[47]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[3819, 11690], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[ ]:
# In[ ]:
# # Environmental Response Genes
# separating Hypo and Hyper
# tldr
#
# In[29]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[30]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[31]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[26]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 11 | sort | uniq -c')
# In[42]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[3210, 10028], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[43]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[3369, 10148], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[47]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[3819, 11690], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[ ]:
# In[ ]:
# # Environmental Response Genes
# separating Hypo and Hyper
# tldr
# 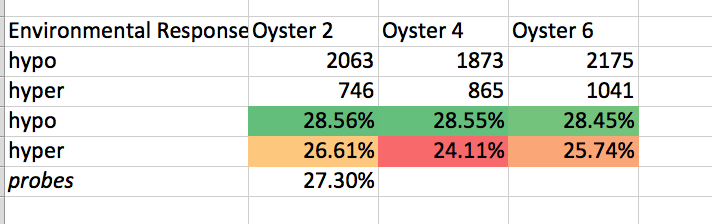 # In[ ]:
# In[33]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[34]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[35]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[27]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 11 | sort | uniq -c')
# In[45]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[2809, 10028], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[48]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[2738, 10148], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[49]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[3216, 11690], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# # TE-Blast
#
# In[ ]:
# In[33]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[34]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[35]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[27]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 11 | sort | uniq -c')
# In[45]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[2809, 10028], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[48]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[2738, 10148], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[49]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[3216, 11690], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# # TE-Blast
# 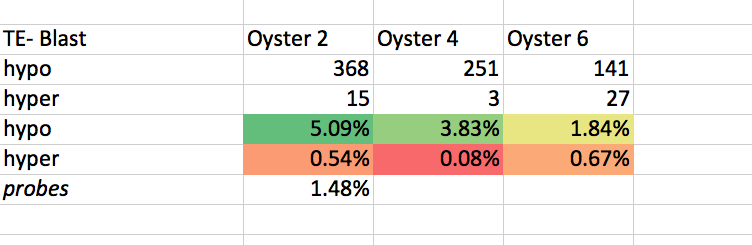 # In[37]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# In[38]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# In[39]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# # Promoter
#
# In[37]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# In[38]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# In[39]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# # Promoter
# 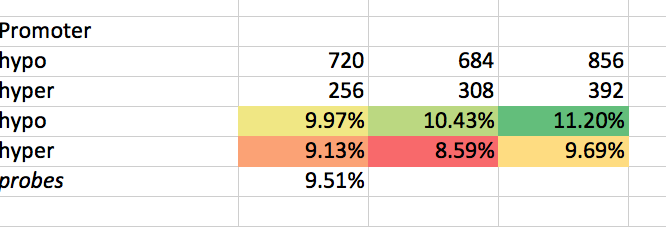 # In[40]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[41]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# !intersectbed \
# -wb \
# -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph \
# -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff \
# | cut -f 6,7 \
# | sort | uniq -c | sed '/#/d'
# !intersectbed \
# -wb \
# -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph \
# -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff \
# | cut -f 6,7 \
# | sort | uniq -c | sed '/#/d'
# # Chi2 test to compare hypo v hyper?
# In[43]:
# Enter the data comparing Oyster ALL hypo versus hyper -HOUSEKEEPING
obs = array([[6527, 21429], [3871, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[44]:
# Enter the data comparing Oyster ALL hypo versus hyper -DEGS
obs = array([[1524, 21429], [692, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[45]:
# Enter the data comparing Oyster ALL hypo versus hyper -TE blast
obs = array([[760, 21429], [45, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
#
# In[40]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[41]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# !intersectbed \
# -wb \
# -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph \
# -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff \
# | cut -f 6,7 \
# | sort | uniq -c | sed '/#/d'
# !intersectbed \
# -wb \
# -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph \
# -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff \
# | cut -f 6,7 \
# | sort | uniq -c | sed '/#/d'
# # Chi2 test to compare hypo v hyper?
# In[43]:
# Enter the data comparing Oyster ALL hypo versus hyper -HOUSEKEEPING
obs = array([[6527, 21429], [3871, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[44]:
# Enter the data comparing Oyster ALL hypo versus hyper -DEGS
obs = array([[1524, 21429], [692, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[45]:
# Enter the data comparing Oyster ALL hypo versus hyper -TE blast
obs = array([[760, 21429], [45, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# 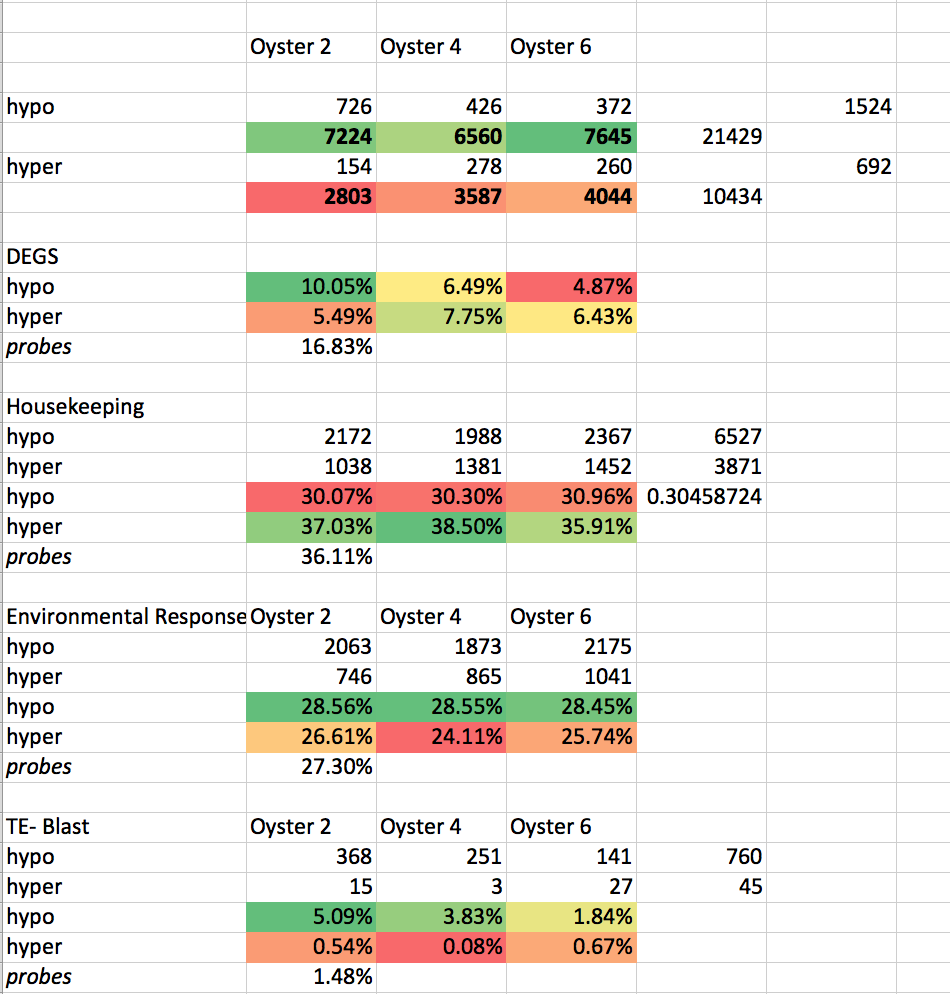 # # Hypo v Hyper on Ensembl gff - gives data on gene body, and repeats
# In[48]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[50]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[49]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[51]:
get_ipython().system("intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 11,12 | sort | uniq -c | sed '/#/d'")
# In[ ]:
# # Hypo v Hyper on Ensembl gff - gives data on gene body, and repeats
# In[48]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[50]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[49]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[51]:
get_ipython().system("intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 11,12 | sort | uniq -c | sed '/#/d'")
# In[ ]:
 # In[2]:
get_ipython().system('date')
# In[20]:
get_ipython().run_line_magic('pylab', 'inline')
import scipy.stats as stats
# Feature (from nb -03)
#
# **tldr** 4 "new" tracks
#
# In[2]:
get_ipython().system('date')
# In[20]:
get_ipython().run_line_magic('pylab', 'inline')
import scipy.stats as stats
# Feature (from nb -03)
#
# **tldr** 4 "new" tracks
#  # ```
# /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf
# /Users/sr320/data-genomic/tentacle/rebuilt.gtf
# /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff
# /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff
# ```
# # DEGs
# `-wb Write the original entry in B for each overlap. Useful for knowing what A overlaps. Restricted by -f and -r.`
# tldr
#
#
# ```
# /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf
# /Users/sr320/data-genomic/tentacle/rebuilt.gtf
# /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff
# /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff
# ```
# # DEGs
# `-wb Write the original entry in B for each overlap. Useful for knowing what A overlaps. Restricted by -f and -r.`
# tldr
#
#  # ## Separating HYPO and HYPER
# In[1]:
get_ipython().system('head ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph')
# In[1]:
get_ipython().system('fgrep -c "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph')
# In[3]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph | head')
# In[4]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph')
# In[6]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph')
# In[7]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph')
# In[ ]:
# In[ ]:
# In[ ]:
# ### HYPO
# In[5]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[9]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[10]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[10]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 11 | sort | uniq -c')
# In[21]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[726, 7224], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[22]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[426, 6560], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[23]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[372, 7645], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# ## HYPER
# In[13]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
# In[12]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
# In[14]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
# In[15]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[16]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[17]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[18]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 11 | sort | uniq -c')
# In[24]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[154, 2803], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[25]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[278, 3587], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[26]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[260, 4044], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[ ]:
# In[ ]:
# In[ ]:
# # Rebuilt (new gtf based on RNAseq data)
# In[15]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[16]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[17]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[18]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 11 | sort | uniq -c')
# In[39]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[8768, 10028], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[40]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[7694, 10148], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[41]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[6160, 11690], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# # Housekeeping Genes
# Separating out hypo and hyper
#
# ## Separating HYPO and HYPER
# In[1]:
get_ipython().system('head ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph')
# In[1]:
get_ipython().system('fgrep -c "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph')
# In[3]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph | head')
# In[4]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph')
# In[6]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph')
# In[7]:
get_ipython().system('fgrep "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph')
# In[ ]:
# In[ ]:
# In[ ]:
# ### HYPO
# In[5]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[9]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[10]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[10]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 11 | sort | uniq -c')
# In[21]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[726, 7224], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[22]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[426, 6560], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[23]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[372, 7645], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# ## HYPER
# In[13]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph')
# In[12]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph')
# In[14]:
get_ipython().system('fgrep -v "-" ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph > /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
get_ipython().system('head /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
get_ipython().system('wc -l /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph')
# In[15]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[16]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[17]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 6 | sort | uniq -c')
# In[18]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cuffdiff_geneexp.sig.gtf | cut -f 11 | sort | uniq -c')
# In[24]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[154, 2803], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[25]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[278, 3587], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[26]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[260, 4044], [117460, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[ ]:
# In[ ]:
# In[ ]:
# # Rebuilt (new gtf based on RNAseq data)
# In[15]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.2M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[16]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.4M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[17]:
get_ipython().system('intersectbed -wb -a ./data/2014.07.02.colson/genomeBrowserTracks/logFC_HS-preHS/2014.07.02.6M_sig.bedGraph -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 6 | sort | uniq -c')
# In[18]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/rebuilt.gtf | cut -f 11 | sort | uniq -c')
# In[39]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[8768, 10028], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[40]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[7694, 10148], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[41]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[6160, 11690], [1197818, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# # Housekeeping Genes
# Separating out hypo and hyper
#  # In[29]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[30]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[31]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[26]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 11 | sort | uniq -c')
# In[42]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[3210, 10028], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[43]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[3369, 10148], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[47]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[3819, 11690], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[ ]:
# In[ ]:
# # Environmental Response Genes
# separating Hypo and Hyper
# tldr
#
# In[29]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[30]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[31]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 6 | sort | uniq -c')
# In[26]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-housekeeping.gff | cut -f 11 | sort | uniq -c')
# In[42]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[3210, 10028], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[43]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[3369, 10148], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[47]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[3819, 11690], [251970, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[ ]:
# In[ ]:
# # Environmental Response Genes
# separating Hypo and Hyper
# tldr
#  # In[ ]:
# In[33]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[34]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[35]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[27]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 11 | sort | uniq -c')
# In[45]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[2809, 10028], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[48]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[2738, 10148], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[49]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[3216, 11690], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# # TE-Blast
#
# In[ ]:
# In[33]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[34]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[35]:
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
get_ipython().system('intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 6 | sort | uniq -c')
# In[27]:
get_ipython().system('intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Users/sr320/data-genomic/tentacle/Cgigas_v9_gene-env-response.gff | cut -f 11 | sort | uniq -c')
# In[45]:
# Enter the data comparing Oyster 2 then Probes
obs = array([[2809, 10028], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[48]:
# Enter the data comparing Oyster 4 then Probes
obs = array([[2738, 10148], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[49]:
# Enter the data comparing Oyster 6 then Probes
obs = array([[3216, 11690], [190475, 697753]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# # TE-Blast
#  # In[37]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# In[38]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# In[39]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# # Promoter
#
# In[37]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# In[38]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# In[39]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_TE-WUBLASTX.gff | cut -f 6 | sort | uniq -c | sed '/#/d'")
# # Promoter
#  # In[40]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[41]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# !intersectbed \
# -wb \
# -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph \
# -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff \
# | cut -f 6,7 \
# | sort | uniq -c | sed '/#/d'
# !intersectbed \
# -wb \
# -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph \
# -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff \
# | cut -f 6,7 \
# | sort | uniq -c | sed '/#/d'
# # Chi2 test to compare hypo v hyper?
# In[43]:
# Enter the data comparing Oyster ALL hypo versus hyper -HOUSEKEEPING
obs = array([[6527, 21429], [3871, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[44]:
# Enter the data comparing Oyster ALL hypo versus hyper -DEGS
obs = array([[1524, 21429], [692, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[45]:
# Enter the data comparing Oyster ALL hypo versus hyper -TE blast
obs = array([[760, 21429], [45, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
#
# In[40]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[41]:
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# !intersectbed \
# -wb \
# -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph \
# -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff \
# | cut -f 6,7 \
# | sort | uniq -c | sed '/#/d'
# !intersectbed \
# -wb \
# -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph \
# -b /Volumes/web-1/trilobite/Crassostrea_gigas_v9_tracks/Cgigas_v9_1k5p_gene_promoter.gff \
# | cut -f 6,7 \
# | sort | uniq -c | sed '/#/d'
# # Chi2 test to compare hypo v hyper?
# In[43]:
# Enter the data comparing Oyster ALL hypo versus hyper -HOUSEKEEPING
obs = array([[6527, 21429], [3871, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[44]:
# Enter the data comparing Oyster ALL hypo versus hyper -DEGS
obs = array([[1524, 21429], [692, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
# In[45]:
# Enter the data comparing Oyster ALL hypo versus hyper -TE blast
obs = array([[760, 21429], [45, 10434]])
# Calculate the chi-square test
chi2_corrected = stats.chi2_contingency(obs, correction=True)
chi2_uncorrected = stats.chi2_contingency(obs, correction=False)
# Print the result
print('CHI SQUARE')
print('The corrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_corrected[0], chi2_corrected[1]))
print('The uncorrected chi2 value is {0:5.3f}, with p={1:5.3f}'.format(chi2_uncorrected[0], chi2_uncorrected[1]))
#  # # Hypo v Hyper on Ensembl gff - gives data on gene body, and repeats
# In[48]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[50]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[49]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[51]:
get_ipython().system("intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 11,12 | sort | uniq -c | sed '/#/d'")
# In[ ]:
# # Hypo v Hyper on Ensembl gff - gives data on gene body, and repeats
# In[48]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.2M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[50]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.4M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[49]:
get_ipython().system('echo "hypo"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hypo.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
get_ipython().system('echo "hyper"')
get_ipython().system("intersectbed -wb -a /Users/sr320/data-genomic/tentacle/2014.07.02.6M_sig.hyper.bedGraph -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 6,7 | sort | uniq -c | sed '/#/d'")
# In[51]:
get_ipython().system("intersectbed -wb -a /Users/sr320/git-repos/paper-Temp-stress/ipynb/data/array-design/OID40453_probe_locations.gff -b /Volumes/web-1/trilobite/Crassostrea_gigas_ensembl_tracks/Crassostrea_gigas.GCA_000297895.1.25.sorted.gff3 | cut -f 11,12 | sort | uniq -c | sed '/#/d'")
# In[ ]: