#!/usr/bin/env python
# coding: utf-8
#
# *This notebook contains course material from [CBE20255](https://jckantor.github.io/CBE20255)
# by Jeffrey Kantor (jeff at nd.edu); the content is available [on Github](https://github.com/jckantor/CBE20255.git).
# The text is released under the [CC-BY-NC-ND-4.0 license](https://creativecommons.org/licenses/by-nc-nd/4.0/legalcode),
# and code is released under the [MIT license](https://opensource.org/licenses/MIT).*
#
# < [Separating Milk](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/03.04-Separating-Milk.ipynb) | [Contents](toc.ipynb) | [Material Balances](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/04.00-Material-Balances.ipynb) > # # Adipic Acid Flowsheet
# ## Summary
#
# This [Jupyter notebook](http://ipython.org/notebook.html) demonstrates the formulation and solution of material balances for a hypothetical adipic acid process described by Murphy (2005, Example 2.15) using the [symbolic algebra package Sympy](http://sympy.org/en/index.html).
# ## Problem Statement
# [Adipic acid](http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=196) (C6H10O4) (also called hexanedioic acid) rarely occurs in nature, but approximately 2.5 billion kilograms are produced per year from petrochemical feedstocks primarily for the production of nylon.
#
#
# # Adipic Acid Flowsheet
# ## Summary
#
# This [Jupyter notebook](http://ipython.org/notebook.html) demonstrates the formulation and solution of material balances for a hypothetical adipic acid process described by Murphy (2005, Example 2.15) using the [symbolic algebra package Sympy](http://sympy.org/en/index.html).
# ## Problem Statement
# [Adipic acid](http://pubchem.ncbi.nlm.nih.gov/summary/summary.cgi?cid=196) (C6H10O4) (also called hexanedioic acid) rarely occurs in nature, but approximately 2.5 billion kilograms are produced per year from petrochemical feedstocks primarily for the production of nylon.
#
# 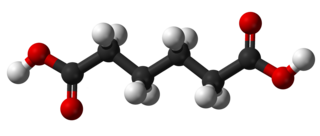 #
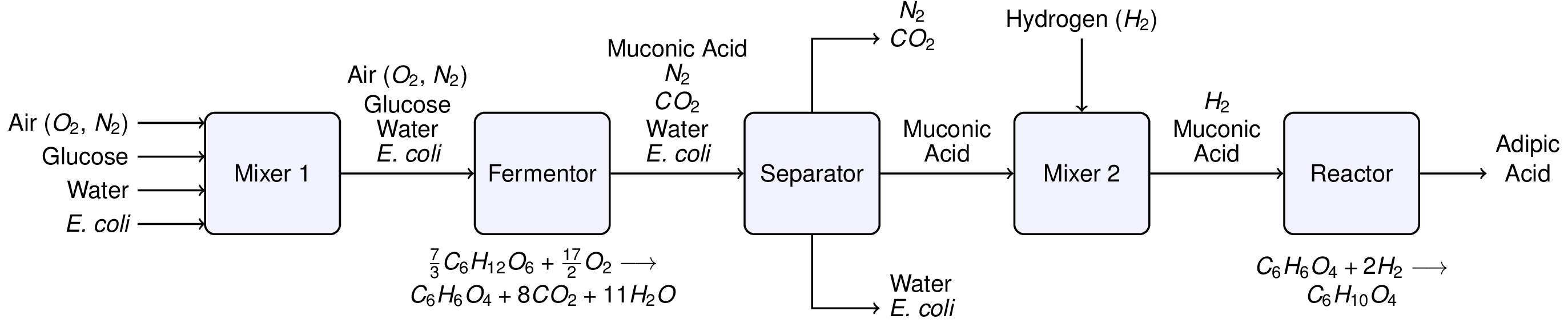
# Recently there has been interest in [producing adipic acid from renewable resources](http://www.ihs.com/products/chemical/technology/pep/bio-based-adipic-acid.aspx). [For example, starting with glucose](http://pubs.acs.org/doi/abs/10.1021/ja00080a057) (C6H12O6), muconic acid (C6H6O4) is produced through fermentation with a genetically modified e. coli. via the reaction
#
# 7⁄3 C6H12O6 + 17⁄2 O2 ➝ C6H6O4 + 8 CO2 + 11 H2O
#
# that is subsequently hydrogentated to form adipic acid
#
# C6H6O4 + 2 H2 ➝ C6H10O4
#
# Murphy (2005, Example 2.15) outlines a hypothetical flowsheet for this process:
#
#
#
# Recently there has been interest in [producing adipic acid from renewable resources](http://www.ihs.com/products/chemical/technology/pep/bio-based-adipic-acid.aspx). [For example, starting with glucose](http://pubs.acs.org/doi/abs/10.1021/ja00080a057) (C6H12O6), muconic acid (C6H6O4) is produced through fermentation with a genetically modified e. coli. via the reaction
#
# 7⁄3 C6H12O6 + 17⁄2 O2 ➝ C6H6O4 + 8 CO2 + 11 H2O
#
# that is subsequently hydrogentated to form adipic acid
#
# C6H6O4 + 2 H2 ➝ C6H10O4
#
# Murphy (2005, Example 2.15) outlines a hypothetical flowsheet for this process:
#
#  #
# Neglecting the _E. coli_, solve for the flowrates necessary to produce 12,000 kg/hour of adipic acid assuming that glucose is available as a 10 mg/mL solution.
#
# ## Solution
# ### Process Variables
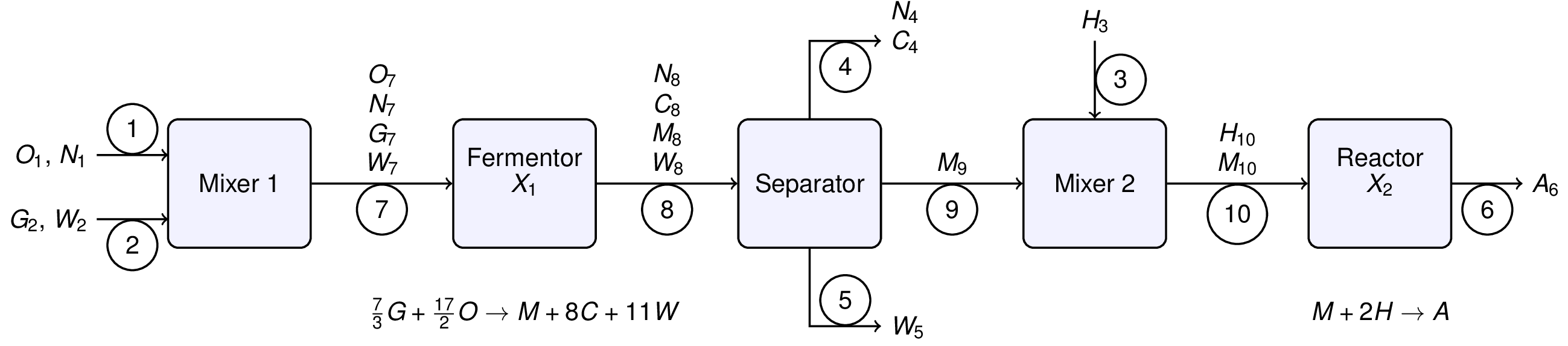
# We begin by relabeling the process flowsheet with stream numbers, stream variables, and extents of reaction. There are chemical species are abbreviated as follows:
#
# * A: adipic acid
# * C: carbon dioxide
# * G: glucose
# * H: hydrogen
# * M: muconic acid
# * N: nitrogen
# * O: oxygen
# * W: water
#
# and where X1 and X2 denote the extents of reactions 1 and 2, respectively. The stream variables denote chemical flowrates in units of kg/hour. The extents of reaction will be in units of kg-mol/hour.
#
#
#
# Neglecting the _E. coli_, solve for the flowrates necessary to produce 12,000 kg/hour of adipic acid assuming that glucose is available as a 10 mg/mL solution.
#
# ## Solution
# ### Process Variables
# We begin by relabeling the process flowsheet with stream numbers, stream variables, and extents of reaction. There are chemical species are abbreviated as follows:
#
# * A: adipic acid
# * C: carbon dioxide
# * G: glucose
# * H: hydrogen
# * M: muconic acid
# * N: nitrogen
# * O: oxygen
# * W: water
#
# and where X1 and X2 denote the extents of reactions 1 and 2, respectively. The stream variables denote chemical flowrates in units of kg/hour. The extents of reaction will be in units of kg-mol/hour.
#
#  # In[1]:
# Import the symbolic algebra package sympy
import sympy as sym
# Extents of reactions 1 and 2
sym.var('X1 X2')
# Stream variables
sym.var('O1 N1') # Stream 1
sym.var('G2 W2') # Stream 2
sym.var('H3') # Stream 3
sym.var('N4 C4') # Stream 4
sym.var('W5') # Stream 5
sym.var('A6') # Stream 6
sym.var('O7 N7 G7 W7') # Stream 7
sym.var('N8 W8 C8 M8') # Stream 8
sym.var('M9') # Stream 9
sym.var('H10 M10') # Stream 10
# Because the flowsheet includes reactions, it will be necessary to include molecular weights in the mass balance expressions. For this purpose we gather the molecular weights of all species into a python dictionary indexed by the chemical species.
# In[2]:
MW = {
'A': 146.14,
'C': 44.01,
'G': 180.16,
'H': 2.02,
'M': 142.11,
'N': 14.01,
'O': 16.00,
'W': 18.02
}
# ### Specifications
# In[3]:
specs = [
sym.Eq(A6, 12000),
sym.Eq(N1/MW['N'], (0.79/0.21)*(O1/MW['O'])),
sym.Eq(G2, 0.01*W2)
]
# ### Material Balances
# In[4]:
mixer1 = [
sym.Eq(0, O1 - O7),
sym.Eq(0, N1 - N7),
sym.Eq(0, G2 - G7),
sym.Eq(0, W2 - W7)
]
reactor1 = [
sym.Eq(0, O7 - MW['O']*(17/2)*X1),
sym.Eq(0, N7 - N8),
sym.Eq(0, G7 - MW['G']*(7/3)*X1),
sym.Eq(0, -C8 + MW['C']*8*X1),
sym.Eq(0, -M8 + MW['M']*X1),
sym.Eq(0, W7 - W8 + MW['W']*11*X1)
]
separator = [
sym.Eq(0, N8 - N4),
sym.Eq(0, C8 - C4),
sym.Eq(0, M8 - M9),
sym.Eq(0, W8 - W5)
]
mixer2 = [
sym.Eq(0, M9 - M10),
sym.Eq(0, H3 - H10)
]
reactor2 = [
sym.Eq(0, H10 - MW['H']*2*X2),
sym.Eq(0, M10 - MW['M']*X2),
sym.Eq(0, -A6 + MW['A']*X2)
]
mbals = mixer1 + reactor1 + separator + mixer2 + reactor2
# In[5]:
specs + mbals
# ### Solution
# In[6]:
soln = sym.solve(mbals + specs)
soln
# In[7]:
for key in soln.keys():
print("{:3s} {:10.1f}".format(str(key), float(soln[key])))
# In[ ]:
#
# < [Separating Milk](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/03.04-Separating-Milk.ipynb) | [Contents](toc.ipynb) | [Material Balances](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/04.00-Material-Balances.ipynb) >
# In[1]:
# Import the symbolic algebra package sympy
import sympy as sym
# Extents of reactions 1 and 2
sym.var('X1 X2')
# Stream variables
sym.var('O1 N1') # Stream 1
sym.var('G2 W2') # Stream 2
sym.var('H3') # Stream 3
sym.var('N4 C4') # Stream 4
sym.var('W5') # Stream 5
sym.var('A6') # Stream 6
sym.var('O7 N7 G7 W7') # Stream 7
sym.var('N8 W8 C8 M8') # Stream 8
sym.var('M9') # Stream 9
sym.var('H10 M10') # Stream 10
# Because the flowsheet includes reactions, it will be necessary to include molecular weights in the mass balance expressions. For this purpose we gather the molecular weights of all species into a python dictionary indexed by the chemical species.
# In[2]:
MW = {
'A': 146.14,
'C': 44.01,
'G': 180.16,
'H': 2.02,
'M': 142.11,
'N': 14.01,
'O': 16.00,
'W': 18.02
}
# ### Specifications
# In[3]:
specs = [
sym.Eq(A6, 12000),
sym.Eq(N1/MW['N'], (0.79/0.21)*(O1/MW['O'])),
sym.Eq(G2, 0.01*W2)
]
# ### Material Balances
# In[4]:
mixer1 = [
sym.Eq(0, O1 - O7),
sym.Eq(0, N1 - N7),
sym.Eq(0, G2 - G7),
sym.Eq(0, W2 - W7)
]
reactor1 = [
sym.Eq(0, O7 - MW['O']*(17/2)*X1),
sym.Eq(0, N7 - N8),
sym.Eq(0, G7 - MW['G']*(7/3)*X1),
sym.Eq(0, -C8 + MW['C']*8*X1),
sym.Eq(0, -M8 + MW['M']*X1),
sym.Eq(0, W7 - W8 + MW['W']*11*X1)
]
separator = [
sym.Eq(0, N8 - N4),
sym.Eq(0, C8 - C4),
sym.Eq(0, M8 - M9),
sym.Eq(0, W8 - W5)
]
mixer2 = [
sym.Eq(0, M9 - M10),
sym.Eq(0, H3 - H10)
]
reactor2 = [
sym.Eq(0, H10 - MW['H']*2*X2),
sym.Eq(0, M10 - MW['M']*X2),
sym.Eq(0, -A6 + MW['A']*X2)
]
mbals = mixer1 + reactor1 + separator + mixer2 + reactor2
# In[5]:
specs + mbals
# ### Solution
# In[6]:
soln = sym.solve(mbals + specs)
soln
# In[7]:
for key in soln.keys():
print("{:3s} {:10.1f}".format(str(key), float(soln[key])))
# In[ ]:
#
# < [Separating Milk](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/03.04-Separating-Milk.ipynb) | [Contents](toc.ipynb) | [Material Balances](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/04.00-Material-Balances.ipynb) >

 #
# Recently there has been interest in [producing adipic acid from renewable resources](http://www.ihs.com/products/chemical/technology/pep/bio-based-adipic-acid.aspx). [For example, starting with glucose](http://pubs.acs.org/doi/abs/10.1021/ja00080a057) (C6H12O6), muconic acid (C6H6O4) is produced through fermentation with a genetically modified e. coli. via the reaction
#
# 7⁄3 C6H12O6 + 17⁄2 O2 ➝ C6H6O4 + 8 CO2 + 11 H2O
#
# that is subsequently hydrogentated to form adipic acid
#
# C6H6O4 + 2 H2 ➝ C6H10O4
#
# Murphy (2005, Example 2.15) outlines a hypothetical flowsheet for this process:
#
#
#
# Recently there has been interest in [producing adipic acid from renewable resources](http://www.ihs.com/products/chemical/technology/pep/bio-based-adipic-acid.aspx). [For example, starting with glucose](http://pubs.acs.org/doi/abs/10.1021/ja00080a057) (C6H12O6), muconic acid (C6H6O4) is produced through fermentation with a genetically modified e. coli. via the reaction
#
# 7⁄3 C6H12O6 + 17⁄2 O2 ➝ C6H6O4 + 8 CO2 + 11 H2O
#
# that is subsequently hydrogentated to form adipic acid
#
# C6H6O4 + 2 H2 ➝ C6H10O4
#
# Murphy (2005, Example 2.15) outlines a hypothetical flowsheet for this process:
#
#  #
# Neglecting the _E. coli_, solve for the flowrates necessary to produce 12,000 kg/hour of adipic acid assuming that glucose is available as a 10 mg/mL solution.
#
# ## Solution
# ### Process Variables
# We begin by relabeling the process flowsheet with stream numbers, stream variables, and extents of reaction. There are chemical species are abbreviated as follows:
#
# * A: adipic acid
# * C: carbon dioxide
# * G: glucose
# * H: hydrogen
# * M: muconic acid
# * N: nitrogen
# * O: oxygen
# * W: water
#
# and where X1 and X2 denote the extents of reactions 1 and 2, respectively. The stream variables denote chemical flowrates in units of kg/hour. The extents of reaction will be in units of kg-mol/hour.
#
#
#
# Neglecting the _E. coli_, solve for the flowrates necessary to produce 12,000 kg/hour of adipic acid assuming that glucose is available as a 10 mg/mL solution.
#
# ## Solution
# ### Process Variables
# We begin by relabeling the process flowsheet with stream numbers, stream variables, and extents of reaction. There are chemical species are abbreviated as follows:
#
# * A: adipic acid
# * C: carbon dioxide
# * G: glucose
# * H: hydrogen
# * M: muconic acid
# * N: nitrogen
# * O: oxygen
# * W: water
#
# and where X1 and X2 denote the extents of reactions 1 and 2, respectively. The stream variables denote chemical flowrates in units of kg/hour. The extents of reaction will be in units of kg-mol/hour.
#
#  # In[1]:
# Import the symbolic algebra package sympy
import sympy as sym
# Extents of reactions 1 and 2
sym.var('X1 X2')
# Stream variables
sym.var('O1 N1') # Stream 1
sym.var('G2 W2') # Stream 2
sym.var('H3') # Stream 3
sym.var('N4 C4') # Stream 4
sym.var('W5') # Stream 5
sym.var('A6') # Stream 6
sym.var('O7 N7 G7 W7') # Stream 7
sym.var('N8 W8 C8 M8') # Stream 8
sym.var('M9') # Stream 9
sym.var('H10 M10') # Stream 10
# Because the flowsheet includes reactions, it will be necessary to include molecular weights in the mass balance expressions. For this purpose we gather the molecular weights of all species into a python dictionary indexed by the chemical species.
# In[2]:
MW = {
'A': 146.14,
'C': 44.01,
'G': 180.16,
'H': 2.02,
'M': 142.11,
'N': 14.01,
'O': 16.00,
'W': 18.02
}
# ### Specifications
# In[3]:
specs = [
sym.Eq(A6, 12000),
sym.Eq(N1/MW['N'], (0.79/0.21)*(O1/MW['O'])),
sym.Eq(G2, 0.01*W2)
]
# ### Material Balances
# In[4]:
mixer1 = [
sym.Eq(0, O1 - O7),
sym.Eq(0, N1 - N7),
sym.Eq(0, G2 - G7),
sym.Eq(0, W2 - W7)
]
reactor1 = [
sym.Eq(0, O7 - MW['O']*(17/2)*X1),
sym.Eq(0, N7 - N8),
sym.Eq(0, G7 - MW['G']*(7/3)*X1),
sym.Eq(0, -C8 + MW['C']*8*X1),
sym.Eq(0, -M8 + MW['M']*X1),
sym.Eq(0, W7 - W8 + MW['W']*11*X1)
]
separator = [
sym.Eq(0, N8 - N4),
sym.Eq(0, C8 - C4),
sym.Eq(0, M8 - M9),
sym.Eq(0, W8 - W5)
]
mixer2 = [
sym.Eq(0, M9 - M10),
sym.Eq(0, H3 - H10)
]
reactor2 = [
sym.Eq(0, H10 - MW['H']*2*X2),
sym.Eq(0, M10 - MW['M']*X2),
sym.Eq(0, -A6 + MW['A']*X2)
]
mbals = mixer1 + reactor1 + separator + mixer2 + reactor2
# In[5]:
specs + mbals
# ### Solution
# In[6]:
soln = sym.solve(mbals + specs)
soln
# In[7]:
for key in soln.keys():
print("{:3s} {:10.1f}".format(str(key), float(soln[key])))
# In[ ]:
#
# < [Separating Milk](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/03.04-Separating-Milk.ipynb) | [Contents](toc.ipynb) | [Material Balances](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/04.00-Material-Balances.ipynb) >
# In[1]:
# Import the symbolic algebra package sympy
import sympy as sym
# Extents of reactions 1 and 2
sym.var('X1 X2')
# Stream variables
sym.var('O1 N1') # Stream 1
sym.var('G2 W2') # Stream 2
sym.var('H3') # Stream 3
sym.var('N4 C4') # Stream 4
sym.var('W5') # Stream 5
sym.var('A6') # Stream 6
sym.var('O7 N7 G7 W7') # Stream 7
sym.var('N8 W8 C8 M8') # Stream 8
sym.var('M9') # Stream 9
sym.var('H10 M10') # Stream 10
# Because the flowsheet includes reactions, it will be necessary to include molecular weights in the mass balance expressions. For this purpose we gather the molecular weights of all species into a python dictionary indexed by the chemical species.
# In[2]:
MW = {
'A': 146.14,
'C': 44.01,
'G': 180.16,
'H': 2.02,
'M': 142.11,
'N': 14.01,
'O': 16.00,
'W': 18.02
}
# ### Specifications
# In[3]:
specs = [
sym.Eq(A6, 12000),
sym.Eq(N1/MW['N'], (0.79/0.21)*(O1/MW['O'])),
sym.Eq(G2, 0.01*W2)
]
# ### Material Balances
# In[4]:
mixer1 = [
sym.Eq(0, O1 - O7),
sym.Eq(0, N1 - N7),
sym.Eq(0, G2 - G7),
sym.Eq(0, W2 - W7)
]
reactor1 = [
sym.Eq(0, O7 - MW['O']*(17/2)*X1),
sym.Eq(0, N7 - N8),
sym.Eq(0, G7 - MW['G']*(7/3)*X1),
sym.Eq(0, -C8 + MW['C']*8*X1),
sym.Eq(0, -M8 + MW['M']*X1),
sym.Eq(0, W7 - W8 + MW['W']*11*X1)
]
separator = [
sym.Eq(0, N8 - N4),
sym.Eq(0, C8 - C4),
sym.Eq(0, M8 - M9),
sym.Eq(0, W8 - W5)
]
mixer2 = [
sym.Eq(0, M9 - M10),
sym.Eq(0, H3 - H10)
]
reactor2 = [
sym.Eq(0, H10 - MW['H']*2*X2),
sym.Eq(0, M10 - MW['M']*X2),
sym.Eq(0, -A6 + MW['A']*X2)
]
mbals = mixer1 + reactor1 + separator + mixer2 + reactor2
# In[5]:
specs + mbals
# ### Solution
# In[6]:
soln = sym.solve(mbals + specs)
soln
# In[7]:
for key in soln.keys():
print("{:3s} {:10.1f}".format(str(key), float(soln[key])))
# In[ ]:
#
# < [Separating Milk](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/03.04-Separating-Milk.ipynb) | [Contents](toc.ipynb) | [Material Balances](http://nbviewer.jupyter.org/github/jckantor/CBE20255/blob/master/notebooks/04.00-Material-Balances.ipynb) >