Vanilla LSTM with CuPy¶
This is a port of Vanilla LSTM with numpy that shows how to run the numpy-based machine learning code on GPU using CuPy. The all contents below is basically copied from the article: Vanilla LSTM with numpy.
This is inspired from Minimal character-level language model with a Vanilla Recurrent Neural Network, in Python/numpy by Andrej Karpathy.
The model usually reaches an error of about 45 after 5000 iterations when tested with 100,000 character sample from Shakespeare. However it sometimes get stuck in a local minima; reinitialize the weights if this happens.
You need to place the input text file as input.txt in the same folder as the python code.
Download data¶
%%bash
curl -L http://cs.stanford.edu/people/karpathy/char-rnn/shakespear.txt -o input.txt
% Total % Received % Xferd Average Speed Time Time Time Current
Dload Upload Total Spent Left Speed
100 99993 100 99993 0 0 70182 0 0:00:01 0:00:01 --:--:-- 149k
Imports¶
import numpy as np
%matplotlib inline
import matplotlib.pyplot as plt
from IPython import display
np.random.seed(2017)
The above imports are from the original article as is. But in this article, you can run almost all computation on GPU by just replacing np with cp. Well, cp is just another name of cupy.
import cupy as cp
cp.random.seed(2017)
To show the difference of computational time by switching CPU and GPU simply, let's use xp instead of np and switch the referenced package between numpy and cupy.
# If you use CPU
# xp = np
# If you use GPU: almost 2 times faster
xp = cp
Because CuPy has high compatibility with NumPy, the same code using NumPy is easily converted for CuPy by just replacing numpy with cupy.
Note:
If the H_size is larger than 1000 or so, training on GPU, namely, using CuPy is several times faster than NumPy. But when the array size is small, the speed gain is often small.
Read and process data¶
data = open('input.txt', 'r').read()
Process data and calculate indexes¶
chars = list(set(data))
data_size, X_size = len(data), len(chars)
print("data has %d characters, %d unique" % (data_size, X_size))
char_to_idx = {ch:i for i,ch in enumerate(chars)}
idx_to_char = {i:ch for i,ch in enumerate(chars)}
data has 99993 characters, 62 unique
Parameters¶
H_size = 100 # Size of the hidden layer
T_steps = 25 # Number of time steps (length of the sequence) used for training
learning_rate = 1e-1 # Learning rate
weight_sd = 0.1 # Standard deviation of weights for initialization
z_size = H_size + X_size # Size of concatenate(H, X) vector
def sigmoid(x):
return 1 / (1 + xp.exp(-x))
def dsigmoid(y):
return y * (1 - y)
def tanh(x):
return xp.tanh(x)
def dtanh(y):
return 1 - y * y
Initialize weights¶
We use random weights with normal distribution (0, weight_sd) for tanh activation function and (0.5, weight_sd) for sigmoid activation function.
Biases are initialized to zeros.
Formulae for LSTM are shown below.
W_f = (xp.random.randn(H_size, z_size) * weight_sd + 0.5).astype(xp.float32)
b_f = xp.zeros((H_size, 1), dtype=xp.float32)
W_i = (xp.random.randn(H_size, z_size) * weight_sd + 0.5).astype(xp.float32)
b_i = xp.zeros((H_size, 1), dtype=xp.float32)
W_C = (xp.random.randn(H_size, z_size) * weight_sd).astype(xp.float32)
b_C = xp.zeros((H_size, 1), dtype=xp.float32)
W_o = (xp.random.randn(H_size, z_size) * weight_sd + 0.5).astype(xp.float32)
b_o = xp.zeros((H_size, 1), dtype=xp.float32)
#For final layer to predict the next character
W_y = (xp.random.randn(X_size, H_size) * weight_sd).astype(xp.float32)
b_y = xp.zeros((X_size, 1))
Gradients¶
dW_f = xp.zeros_like(W_f, dtype=xp.float32)
dW_i = xp.zeros_like(W_i, dtype=xp.float32)
dW_C = xp.zeros_like(W_C, dtype=xp.float32)
dW_o = xp.zeros_like(W_o, dtype=xp.float32)
dW_y = xp.zeros_like(W_y, dtype=xp.float32)
db_f = xp.zeros_like(b_f, dtype=xp.float32)
db_i = xp.zeros_like(b_i, dtype=xp.float32)
db_C = xp.zeros_like(b_C, dtype=xp.float32)
db_o = xp.zeros_like(b_o, dtype=xp.float32)
db_y = xp.zeros_like(b_y, dtype=xp.float32)
Forward pass¶
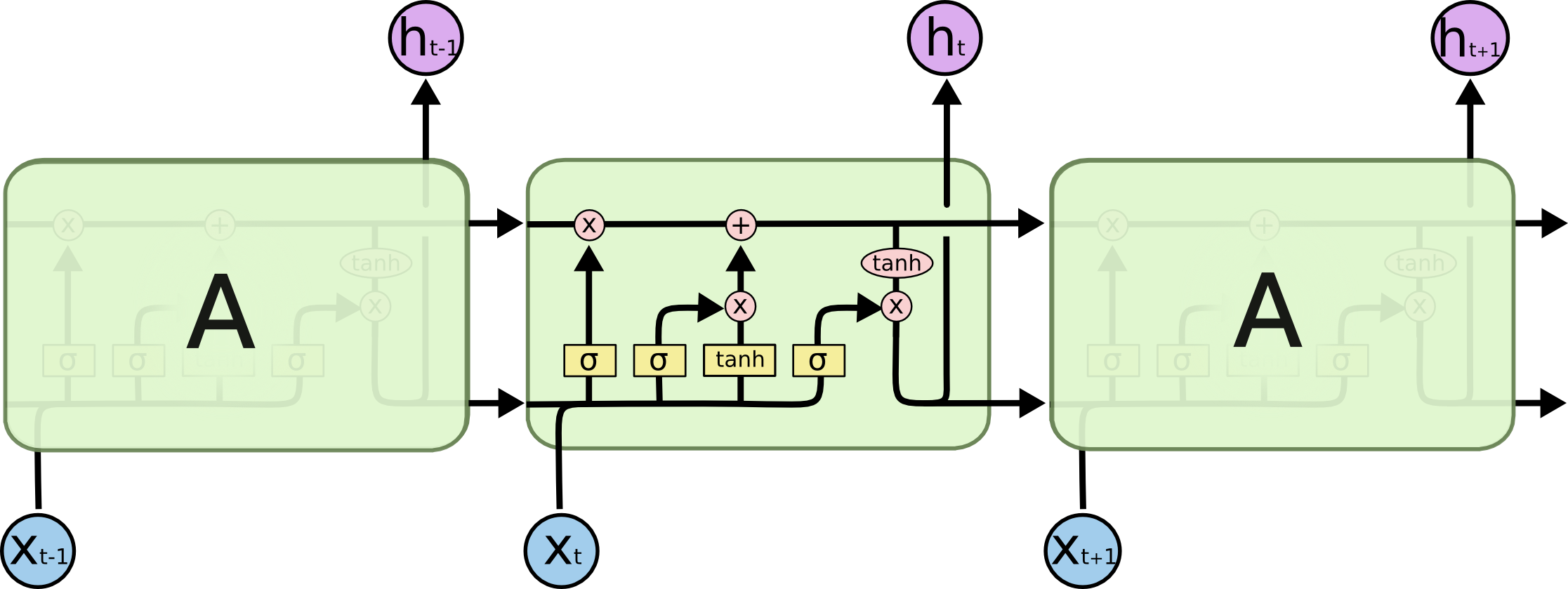
Image taken from Understanding LSTM Networks. Please read the article for a good explanation of LSTMs.
Concatenation of $h_{t-1}$ and $x_t$¶
$$ z = [h_{t-1}, x_t] $$LSTM functions¶
$$ \begin{eqnarray} f_t &=& \sigma(W_f \cdot z + b_f) \\ i_t &=& \sigma(W_i \cdot z + b_i) \\ \bar{C}_t &=& \tanh(W_C \cdot z + b_C) \\ C_t &=& f_t \ast C_{t-1} + i_t \ast \bar{C}_t \\ o_t &=& \sigma(W_O \cdot z + b_t) \\ h_t &=& o_t \ast \tanh(C_t) \end{eqnarray} $$Logits¶
$$ y_t = W_y \cdot h_t + b_y $$Softmax¶
$$ \hat{p}_t = {\rm softmax}(y_t) $$$\hat{p}_t$ is p in code and $p_t$ is targets.
def forward(x, h_prev, C_prev):
assert x.shape == (X_size, 1)
assert h_prev.shape == (H_size, 1)
assert C_prev.shape == (H_size, 1)
z = xp.concatenate((h_prev, x))
f = sigmoid(xp.dot(W_f, z) + b_f)
i = sigmoid(xp.dot(W_i, z) + b_i)
C_bar = tanh(xp.dot(W_C, z) + b_C)
C = f * C_prev + i * C_bar
o = sigmoid(xp.dot(W_o, z) + b_o)
h = o * tanh(C)
y = xp.dot(W_y, h) + b_y
y -= y.max()
p = xp.exp(y) / xp.sum(xp.exp(y))
return z, f, i, C_bar, C, o, h, y, p
Backward pass¶
Loss¶
$$ \mathcal{L} = - \sum p_{t,j}log \hat{p}_{t,j} $$Gradients¶
$$ \begin{eqnarray} dy_t &=& \hat{p}_t - p_t \\ dh_t &=& dh'_{t+1} + W_y^T \cdot d_y \\ do_t &=& dh_t \ast \tanh (C_t) \\ dC_t &=& dC'_{t+q} + dh_t \ast o_t \ast (1 - \tanh^2(C_t)) \\ d \bar{C}_t &=& d C_t \ast i_t \\ d i_t &=& d C_t \ast \bar{C}_t \\ d f_t &=& d C_t \ast C_{t-1} \\ d z_t &=& W_f^T \cdot df_t + W_i^T \cdot di_t + W_C^T \cdot \bar{C}_t + W^T_o \cdot do_t \\ [dh'_t, dx_t] &=& dz_t \\ dC'_t &=& f \ast dC_t \end{eqnarray} $$targetis target character index $p_t$dh_nextis $dh_{t+1}$ (size $H \times 1$)dC_nextis $dC_{t+1}$ (size $H \times 1$)C_previs $C_{t-1}$ (size $H \times 1$)- Returns $dh_t$ and $dC_t$
def backward(target, dh_next, dC_next, C_prev, z, f, i, C_bar, C, o, h, y, p):
global dW_f, dW_i, dW_C, dW_o, dW_y
global db_f, db_i, db_C, db_o, db_y
assert z.shape == (X_size + H_size, 1)
assert y.shape == (X_size, 1)
assert p.shape == (X_size, 1)
for param in [dh_next, dC_next, C_prev, f, i, C_bar, C, o, h]:
assert param.shape == (H_size, 1)
dy = xp.copy(p)
dy[target] -= 1
dW_y += xp.dot(dy, h.T)
db_y += dy
dh = xp.dot(W_y.T, dy)
dh += dh_next
do = dh * tanh(C)
do = dsigmoid(o) * do
dW_o += xp.dot(do, z.T)
db_o += do
dC = xp.copy(dC_next)
dC += dh * o * dtanh(tanh(C))
dC_bar = dC * i
dC_bar = dC_bar * dtanh(C_bar)
dW_C += xp.dot(dC_bar, z.T)
db_C += dC_bar
di = dC * C_bar
di = dsigmoid(i) * di
dW_i += xp.dot(di, z.T)
db_i += di
df = dC * C_prev
df = dsigmoid(f) * df
dW_f += xp.dot(df, z.T)
db_f += df
dz = xp.dot(W_f.T, df) \
+ xp.dot(W_i.T, di) \
+ xp.dot(W_C.T, dC_bar) \
+ xp.dot(W_o.T, do)
dh_prev = dz[:H_size, :]
dC_prev = f * dC
return dh_prev, dC_prev
Forward Backward Pass¶
Calculate and store the values in forward pass. Accumulate gradients in backward pass and clip gradients to avoid exploding gradients.
input,targetare list of integers, with character indexes.h_previs the array of initialhat $h_1$ (size $H \times 1$)C_previs the array of initialCat $C_1$ (size $H \times 1$)- Returns loss, final $h_T$ and $C_T$
def forward_backward(inputs, targets, h_prev, C_prev):
# To store the values for each time step
x_s, z_s, f_s, i_s, C_bar_s, C_s, o_s, h_s, y_s, p_s = {}, {}, {}, {}, {}, {}, {}, {}, {}, {}
# Values at t - 1
h_s[-1] = xp.copy(h_prev)
C_s[-1] = xp.copy(C_prev)
loss = 0
# Loop through time steps
assert len(inputs) == T_steps
for t in range(len(inputs)):
x_s[t] = xp.zeros((X_size, 1), dtype=xp.float32)
x_s[t][inputs[t]] = 1 # Input character
z_s[t], f_s[t], i_s[t], C_bar_s[t], C_s[t], o_s[t], h_s[t], y_s[t], p_s[t] \
= forward(x_s[t], h_s[t - 1], C_s[t - 1]) # Forward pass
loss += -xp.log(p_s[t][targets[t], 0]) # Loss for at t
for dparam in [dW_f, dW_i, dW_C, dW_o, dW_y, db_f, db_i, db_C, db_o, db_y]:
dparam.fill(0)
dh_next = xp.zeros_like(h_s[0], dtype=xp.float32) #dh from the next character
dC_next = xp.zeros_like(C_s[0], dtype=xp.float32) #dh from the next character
for t in reversed(range(len(inputs))):
# Backward pass
dh_next, dC_next = backward(target = targets[t], dh_next = dh_next, dC_next = dC_next, C_prev = C_s[t-1],
z = z_s[t], f = f_s[t], i = i_s[t], C_bar = C_bar_s[t], C = C_s[t], o = o_s[t],
h = h_s[t], y = y_s[t], p = p_s[t])
# Clip gradients to mitigate exploding gradients
for dparam in [dW_f, dW_i, dW_C, dW_o, dW_y, db_f, db_i, db_C, db_o, db_y]:
xp.clip(dparam, -1, 1, out=dparam)
return loss, h_s[len(inputs) - 1], C_s[len(inputs) - 1]
Sample the next character¶
def sample(h_prev, C_prev, first_char_idx, sentence_length):
x = xp.zeros((X_size, 1))
x[first_char_idx] = 1
h = h_prev
C = C_prev
indexes = []
for t in range(sentence_length):
_, _, _, _, C, _, h, _, p = forward(x, h, C)
assert xp.all(p.ravel() >= 0)
idx = xp.random.choice(xp.arange(X_size), size=(1,), p=p.ravel())[0]
x = xp.zeros((X_size, 1), dtype=xp.float32)
x[idx] = 1
indexes.append(int(idx))
return indexes
Training (Adagrad)¶
$$ w = w - \eta \frac{dw}{\sum dw^2_{\tau}} $$def update_status(inputs, h_prev, C_prev):
#initialized later
global plot_iter, plot_loss
global smooth_loss
# Get predictions for 200 letters with current model
display.clear_output(wait=True)
sample_idx = sample(h_prev, C_prev, inputs[0], 200)
txt = ''.join(idx_to_char[idx] for idx in sample_idx)
# Clear and plot
plt.clf()
plt.plot(plot_iter, plot_loss)
display.display(plt.gcf())
#Print prediction and loss
print("----\n %s \n----" % (txt, ))
print("iter %d, loss %f" % (iteration, smooth_loss))
Memory variables for Adagrad
mW_f = xp.zeros_like(W_f, dtype=xp.float32)
mW_i = xp.zeros_like(W_i, dtype=xp.float32)
mW_C = xp.zeros_like(W_C, dtype=xp.float32)
mW_o = xp.zeros_like(W_o, dtype=xp.float32)
mW_y = xp.zeros_like(W_y, dtype=xp.float32)
mb_f = xp.zeros_like(b_f, dtype=xp.float32)
mb_i = xp.zeros_like(b_i, dtype=xp.float32)
mb_C = xp.zeros_like(b_C, dtype=xp.float32)
mb_o = xp.zeros_like(b_o, dtype=xp.float32)
mb_y = xp.zeros_like(b_y, dtype=xp.float32)
# Exponential average of loss
# Initialize to a error of a random model
smooth_loss = (-xp.log(1.0 / X_size) * T_steps).astype(xp.float32)
iteration, p = 0, 0
# For the graph
plot_iter = np.zeros((0))
plot_loss = np.zeros((0))
%%time
while True:
# Try catch for interruption
try:
# Reset
if p + T_steps >= len(data) or iteration == 0:
g_h_prev = xp.zeros((H_size, 1), dtype=xp.float32)
g_C_prev = xp.zeros((H_size, 1), dtype=xp.float32)
p = 0
inputs = [char_to_idx[ch] for ch in data[p: p + T_steps]]
targets = [char_to_idx[ch] for ch in data[p + 1: p + T_steps + 1]]
loss, g_h_prev, g_C_prev = forward_backward(inputs, targets, g_h_prev, g_C_prev)
smooth_loss = smooth_loss * 0.999 + loss * 0.001
# Print every hundred steps
if iteration % 100 == 0:
update_status(inputs, g_h_prev, g_C_prev)
# Update weights
for param, dparam, mem in zip([W_f, W_i, W_C, W_o, W_y, b_f, b_i, b_C, b_o, b_y],
[dW_f, dW_i, dW_C, dW_o, dW_y, db_f, db_i, db_C, db_o, db_y],
[mW_f, mW_i, mW_C, mW_o, mW_y, mb_f, mb_i, mb_C, mb_o, mb_y]):
mem += dparam * dparam # Calculate sum of gradients
#print(learning_rate * dparam)
param += -(learning_rate * dparam / xp.sqrt(mem + 1e-8))
plot_iter = np.append(plot_iter, [iteration])
if isinstance(loss, cp.ndarray):
loss = loss.get()
plot_loss = np.append(plot_loss, [loss])
p += T_steps
iteration += 1
if iteration > 6000:
break
except KeyboardInterrupt:
update_status(inputs, g_h_prev, g_C_prev)
break
---- outhant her abnuck a casthantes Pandors, Are you corpore; surt is. in Is boo all you KLUS: Goke to knowt but, he have be pedaen lost'd Cold led. Ravert was, soperen, but brous. KIN: -flak Thym;, Wit ---- iter 6000, loss 49.692995 CPU times: user 12min 4s, sys: 15.6 s, total: 12min 19s Wall time: 12min 21s
Gradient Check¶
Approximate the numerical gradients by changing parameters and running the model. Check if the approximated gradients are equal to the computed analytical gradients (by backpropagation).
Try this on num_checks individual paramters picked randomly for each weight matrix and bias vector.
from random import uniform
def gradient_check(inputs, target, h_prev, C_prev):
global W_f, W_i, W_C, W_o, W_y, b_f, b_i, b_C, b_o, b_y
global dW_f, dW_i, dW_C, dW_o, dW_y, db_f, db_i, db_C, db_o, db_y
num_checks = 10 # Number of parameters to test
delta = 1e-5 # The change to make on the parameter
# To calculate computed gradients
_, _, _ = forward_backward(inputs, targets, h_prev, C_prev)
for param, dparam, name in zip([W_f, W_i, W_C, W_o, W_y, b_f, b_i, b_C, b_o, b_y],
[dW_f, dW_i, dW_C, dW_o, dW_y, db_f, db_i, db_C, db_o, db_y],
['W_f', 'W_i', 'W_C', 'W_o', 'W_y', 'b_f', 'b_i', 'b_C', 'b_o', 'b_y']):
assert param.shape == dparam.shape
dparam_copy = xp.copy(dparam) #Make a copy because this will get modified
# Test num_checks times
for i in range(num_checks):
# Pick a random index
rnd_idx = int(uniform(0,param.size))
# evaluate cost at [x + delta] and [x - delta]
old_val = xp.ravel(param)[rnd_idx]
xp.ravel(param)[rnd_idx] = old_val + delta
loss_plus_delta, _, _ = forward_backward(inputs, targets, h_prev, C_prev)
xp.ravel(param)[rnd_idx] = old_val - delta
loss_mins_delta, _, _ = forward_backward(inputs, targets, h_prev, C_prev)
xp.ravel(param)[rnd_idx] = old_val
grad_analytical = xp.ravel(dparam_copy)[rnd_idx]
grad_numerical = (loss_plus_delta - loss_mins_delta) / (2 * delta)
# Clip numerical error because grad_analytical is clipped
grad_numerical = xp.clip(grad_numerical, -1, 1)
err_sum = abs(grad_numerical + grad_analytical) + 1e-09
rel_error = abs(grad_analytical - grad_numerical) / err_sum
# If relative error is greater than 1e-06
if rel_error > 1e-06:
print('%s (%e, %e) => %e' % (name, grad_numerical, grad_analytical, rel_error))
gradient_check(inputs, targets, g_h_prev, g_C_prev)
W_f (-5.788818e-03, -1.894525e-03) => 5.068488e-01 W_f (-4.424725e-03, -2.570756e-03) => 2.650237e-01 W_f (-1.574892e-02, 3.109146e-04) => 1.040279e+00 W_f (2.522772e-02, -2.049634e-05) => 1.001626e+00 W_f (-2.772584e-02, -3.596031e-04) => 9.743922e-01 W_f (-9.696586e-02, -3.551726e-02) => 4.638221e-01 W_f (-1.919746e-02, -4.452238e-05) => 9.953723e-01 W_i (-2.128421e-02, -1.987336e-03) => 8.292046e-01 W_i (-9.618537e-03, -1.770927e-03) => 6.890236e-01 W_i (-5.882980e-02, 1.477409e-03) => 1.051520e+00 W_i (-4.811230e-02, -8.973669e-03) => 6.856085e-01 W_i (-2.069689e-02, 2.397710e-03) => 1.262057e+00 W_C (-9.150008e-03, -4.846476e-03) => 3.074723e-01 W_C (-5.384253e-02, 3.501519e-03) => 1.139112e+00 W_C (0.000000e+00, 1.724286e-05) => 9.999420e-01 W_C (-3.551228e-02, -4.914250e-03) => 7.568799e-01 W_C (0.000000e+00, 2.751392e-05) => 9.999637e-01 W_C (-2.206686e-02, 1.656087e-04) => 1.015123e+00 W_C (-4.734427e-03, 3.258538e-02) => 1.339983e+00 W_C (5.200395e-03, 4.812866e-02) => 8.049695e-01 W_o (1.664602e-02, 1.216984e-02) => 1.553371e-01 W_o (-3.094253e-03, -1.786075e-02) => 7.046765e-01 W_y (5.069651e-05, 9.411758e-05) => 2.998380e-01 W_y (1.354294e-06, 3.190190e-06) => 4.038944e-01 W_y (-1.707993e-01, -3.533963e-01) => 3.483377e-01 W_y (-3.616201e-05, -6.439822e-05) => 2.807863e-01 W_y (1.045780e-04, 2.551565e-04) => 4.185810e-01 W_y (-7.008616e-05, -6.961373e-05) => 3.381729e-03 W_y (-1.320173e-03, -3.049548e-03) => 3.957632e-01 W_y (-2.135536e-05, -5.128610e-05) => 4.120281e-01 W_y (-3.736993e-05, -3.475788e-05) => 3.621374e-02 W_y (4.692321e-03, 1.760141e-02) => 5.790457e-01 b_f (9.625265e-02, 1.281972e-01) => 1.423236e-01 b_f (-1.711278e-02, -8.734714e-03) => 3.241346e-01 b_f (-6.363861e-02, -9.571880e-02) => 2.013097e-01 b_f (-1.353493e-01, -2.709278e-01) => 3.337093e-01 b_f (1.295448e-01, 2.294842e-01) => 2.783604e-01 b_f (0.000000e+00, 1.190781e-05) => 9.999160e-01 b_f (-1.097183e-02, 2.446750e-02) => 2.625979e+00 b_f (6.660086e-03, 1.548321e-01) => 9.175182e-01 b_f (-3.765070e-02, -1.929383e-03) => 9.025073e-01 b_f (8.172772e-03, -1.733225e-02) => 2.784549e+00 b_i (-1.797925e-02, 3.960431e-03) => 1.565016e+00 b_i (2.327358e-04, 1.566012e-02) => 9.707118e-01 b_i (3.910756e-05, 1.577477e-02) => 9.950540e-01 b_i (-1.699091e-01, -2.288668e-01) => 1.478467e-01 b_i (-7.337276e-02, -9.197736e-02) => 1.125164e-01 b_i (-3.058013e-02, 9.644181e-03) => 1.921303e+00 b_i (6.721953e-02, 1.486464e-01) => 3.772104e-01 b_i (-8.904879e-02, -5.259708e-02) => 2.573440e-01 b_i (-8.196457e-02, -5.133832e-02) => 2.297493e-01 b_i (-1.127150e-02, 2.934633e-03) => 1.704013e+00 b_C (-8.878277e-02, -9.282753e-02) => 2.227166e-02 b_C (-1.053689e-01, -2.198471e-01) => 3.520066e-01 b_C (-1.254594e-01, -1.306037e-01) => 2.008990e-02 b_C (-8.069183e-03, 4.495833e-02) => 1.437483e+00 b_C (1.315191e-01, 3.286199e-01) => 4.283507e-01 b_C (-2.857717e-01, -5.199677e-01) => 2.906598e-01 b_C (-3.367491e-01, -5.167727e-01) => 2.109186e-01 b_C (-8.336149e-02, -2.002720e-01) => 4.121887e-01 b_C (-2.030515e-01, -2.748539e-01) => 1.502439e-01 b_C (-2.615591e-01, -5.192239e-01) => 3.300082e-01 b_o (-1.828830e-01, -3.566602e-01) => 3.220820e-01 b_o (-6.164580e-02, -6.660104e-02) => 3.863832e-02 b_o (3.400145e-03, 4.486300e-02) => 8.590997e-01 b_o (-8.065856e-02, 1.755721e-02) => 1.556476e+00 b_o (-3.057445e-02, -1.059484e-02) => 4.853036e-01 b_o (-6.164580e-02, -6.660104e-02) => 3.863832e-02 b_o (-7.878454e-03, 8.931704e-03) => 1.596026e+01 b_o (-3.220456e-02, 8.119028e-03) => 1.674183e+00 b_o (-1.044374e-01, -1.323533e-01) => 1.178925e-01 b_o (-1.055425e-01, -1.247781e-01) => 8.351679e-02 b_y (4.384235e-02, 8.768427e-02) => 3.333312e-01 b_y (7.502261e-03, 1.500445e-02) => 3.333311e-01 b_y (-1.380362e-01, -2.760797e-01) => 3.333449e-01 b_y (2.693329e-01, 5.386631e-01) => 3.333312e-01 b_y (3.615790e-03, 7.231543e-03) => 3.333311e-01 b_y (-2.748632e-01, -5.497285e-01) => 3.333350e-01 b_y (-1.380362e-01, -2.760797e-01) => 3.333449e-01 b_y (-1.380362e-01, -2.760797e-01) => 3.333449e-01 b_y (8.233201e-05, 1.646628e-04) => 3.333287e-01 b_y (8.127466e-02, 1.625486e-01) => 3.333313e-01