Python for Data Scientists
*v1.0 Last updated April 2, 2018*
# Python Basics
Data Types¶
You can check an object's type using the type() built-in function.
Integers¶
Let's take a look at the type() of the integer from above:
# Print type() of 2
print(type(2))
<class 'int'>
Truncations
The int() function creats integers. It always rounds down. That's because what it's really doing is truncating the decimals from the number.
print( int(5.3) )
print( int(5.9999) )
5 5
Floats¶
Floats are numeric objects with decimals:
print(type(5))
print(type(5.))
print(type(5.3))
<class 'int'> <class 'float'> <class 'float'>
You can also convert an integer into a float using the float() function. Python will tag on a .0 to indicate that it's a float.
print( 1337 ) # int
print( float(1337) ) # convert int to float
1337 1337.0
Strings¶
Strings are text objects enclosed by single or double quotes.
print( 'This is a string.' )
print( type('This is a string.') )
This is a string. <class 'str'>
Calculations¶
Operators¶
The following are the basic operations. Here is a full list fo perators.
print( 10 + 2 ) # Add
print( 10 - 2 ) # Subtract
print( 10 / 2 ) # Divide
print( 10 * 2 ) # Multiply
print( 10**2 ) # Squared
print( 10 % 2 ) # Modulo (i.e. what is the remainder when 10 is divided by 2?)
12 8 5.0 20 100 0
Comparison operators¶
Comparison operators are often based on boolean logic. These are binary obeject types that are either True or False.
print(5>3) # Greater than
print(5>=3) # Greater than or equal to
print(5< 3) # Less than
print(5<= 3) # Less than or equal to
print(5== 3) # Equal to
print(5!= 3) # Not equal to
True True False False False True
Example string opeartions
'5' * 5 # Repetition
'55555'
print( 992 + 345 ) # Performs calculation
print( '992 + 345' ) # Does not perform calculation
1337 992 + 345
'992' + '345' # Concatentation
'992345'
Analysis: Calculating volume¶
This is the start of a long series of connected analysis seen throughout the notebook. Each analysis build on previous analyses and concepts.
Here is our hypothetical setting:
- You are 007 James Bond. You are on your first training mission.
- You will travel to different locations in London.
- First, you will prepare for and plan the trip.
- After you arrive at each location, you will meet with local business owners and offer your help.
First, calculate the total volume of water you can carry, in cm$^3$.
- The formula for the volume of a cylinder is $V = \pi r^2 h$
- The container cylinder's radius is 4cm and its height is 16cm
- You have room in your backpack for 3 containers!
# Total volume of water you can carry
import math # need the pi constant
print(3 * 3.14 * 4**2 * 16)
3 * math.pi * 4**2 * 16
2411.52
2412.743157956961
Q estimates that you'll need at least 2000 cm$^3$ for the mission. Is our container large enough?
(3 * math.pi * 4**2 * 16) > 2000
True
Formatting¶
# Insert dynamic value into string
print( '1 + 2 = {}'.format(1 + 2) )
1 + 2 = 3
# Insert 3 dynamic values into string
print( '{} + {} = {}'.format(1, 2, 1 + 2) )
1 + 2 = 3
Variables¶
Variables are named objects and used to store data or other information:
# Set variables
a = 2
b = 4
c = "Hello from Canada"
print( a )
print( b )
print( c )
print( a * b )
2 4 Hello from Canada 8
Naming conventions¶
Best practice: use lower_case_with_underscores. The shorter the variable the better.
For example, let's say we wanted to calculate total return on investment based on the formula for compound interest:
# Set variables
principal = 1000.0
yearly_interest_rate = 0.03
n_years = 5
# Calculate total investment return
total_return = principal * (1 + yearly_interest_rate)**n_years - principal
# Print total investment return
print( total_return )
159.27407430000017
Interlude: Getting help on python commands
# Get help on the print() function
help(print)
Help on built-in function print in module builtins:
print(...)
print(value, ..., sep=' ', end='\n', file=sys.stdout, flush=False)
Prints the values to a stream, or to sys.stdout by default.
Optional keyword arguments:
file: a file-like object (stream); defaults to the current sys.stdout.
sep: string inserted between values, default a space.
end: string appended after the last value, default a newline.
flush: whether to forcibly flush the stream.
# Data Structures
Lists¶
Lists are mutable sequences of objects, enclosed by square brackets: []
# Create list of integers
integer_list = [0, 1, 2, 3, 4]
print( integer_list )
print( type(integer_list) )
[0, 1, 2, 3, 4] <class 'list'>
# Create list of mixed types: strings, ints, and floats
my_list = ['hello', 1, 'Canada', 2, 3.0]
# Print entire list
print( my_list )
['hello', 1, 'Canada', 2, 3.0]
Indexing lists¶
In Python, lists are zero-indexed.
print( my_list[0] ) # Print the first element
print( my_list[2] ) # Print the third element
hello Canada
Slicing
#Selects all starting from the 2nd element, but BEFORE the 4th element
print( my_list[1:3] )
[1, 'Canada']
# Selects all BEFORE the 4th element
print( my_list[:3] )
# Selects all starting from the 2nd element
print( my_list[1:] )
['hello', 1, 'Canada'] [1, 'Canada', 2, 3.0]
Negative indices
# Select the last element
print( my_list[-1] )
# Select all BEFORE the last element
print( my_list[:-1] )
# Selects all starting from the 2nd element, but before the last element
print( my_list[1:-1] )
3.0 ['hello', 1, 'Canada', 2] [1, 'Canada', 2]
Mutable¶
Lists are mutable, meaning you can change individual elements of a list. Lets update my_list:
my_list[0] = 'bonjour' # Sets new value for the first element
print( my_list[0] ) # Print the first element
print( my_list[2] ) # Print the third element
bonjour Canada
Appending to and removing elements from lists are both easy to do. These are a special class of 'list' functions called methods. You can see all list methods by calling the help for lists!
help(list)
Help on class list in module builtins: class list(object) | list() -> new empty list | list(iterable) -> new list initialized from iterable's items | | Methods defined here: | | __add__(self, value, /) | Return self+value. | | __contains__(self, key, /) | Return key in self. | | __delitem__(self, key, /) | Delete self[key]. | | __eq__(self, value, /) | Return self==value. | | __ge__(self, value, /) | Return self>=value. | | __getattribute__(self, name, /) | Return getattr(self, name). | | __getitem__(...) | x.__getitem__(y) <==> x[y] | | __gt__(self, value, /) | Return self>value. | | __iadd__(self, value, /) | Implement self+=value. | | __imul__(self, value, /) | Implement self*=value. | | __init__(self, /, *args, **kwargs) | Initialize self. See help(type(self)) for accurate signature. | | __iter__(self, /) | Implement iter(self). | | __le__(self, value, /) | Return self<=value. | | __len__(self, /) | Return len(self). | | __lt__(self, value, /) | Return self<value. | | __mul__(self, value, /) | Return self*value.n | | __ne__(self, value, /) | Return self!=value. | | __new__(*args, **kwargs) from builtins.type | Create and return a new object. See help(type) for accurate signature. | | __repr__(self, /) | Return repr(self). | | __reversed__(...) | L.__reversed__() -- return a reverse iterator over the list | | __rmul__(self, value, /) | Return self*value. | | __setitem__(self, key, value, /) | Set self[key] to value. | | __sizeof__(...) | L.__sizeof__() -- size of L in memory, in bytes | | append(...) | L.append(object) -> None -- append object to end | | clear(...) | L.clear() -> None -- remove all items from L | | copy(...) | L.copy() -> list -- a shallow copy of L | | count(...) | L.count(value) -> integer -- return number of occurrences of value | | extend(...) | L.extend(iterable) -> None -- extend list by appending elements from the iterable | | index(...) | L.index(value, [start, [stop]]) -> integer -- return first index of value. | Raises ValueError if the value is not present. | | insert(...) | L.insert(index, object) -- insert object before index | | pop(...) | L.pop([index]) -> item -- remove and return item at index (default last). | Raises IndexError if list is empty or index is out of range. | | remove(...) | L.remove(value) -> None -- remove first occurrence of value. | Raises ValueError if the value is not present. | | reverse(...) | L.reverse() -- reverse *IN PLACE* | | sort(...) | L.sort(key=None, reverse=False) -> None -- stable sort *IN PLACE* | | ---------------------------------------------------------------------- | Data and other attributes defined here: | | __hash__ = None
# Add to end of the list
my_list.append(22)
print(my_list)
# Remove an element from the list
my_list.remove(3.0)
print(my_list)
['bonjour', 1, 'Canada', 2, 3.0, 22] ['bonjour', 1, 'Canada', 2, 22]
List Operations¶
Lets create a couple of lists and do some operations.
a = [1, 2, 3]
b = [4, 5, 6]
print( a + b ) # Concatentation
[1, 2, 3, 4, 5, 6]
print( a * 3 ) # Repetition
[1, 2, 3, 1, 2, 3, 1, 2, 3]
print( 3 in a ) # Membership
True
print( min(b), max(b) ) # Min, Max
4 6
print( len(a) ) # Length
3
Analysis: Reading in data and creating variables¶
In previous analysis, you learned about 007's training mission. You also made sure you had enough water for the trip by performing calculations. Now, it's time to start planning the locations in London to visit!
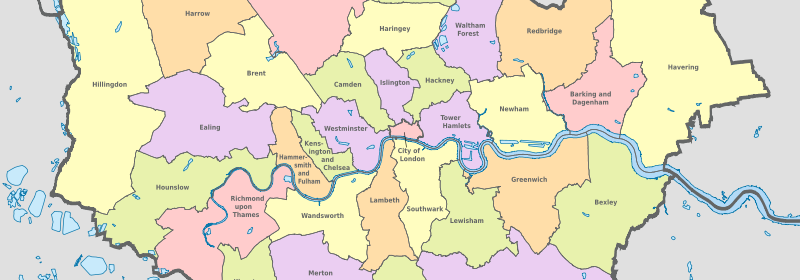
Boroughs of London.
Lets analayze some London boroughs and each of there locations. First. lets create lists of the neccessary borough variables that contain the locations. Python has a variety of input/output methods. If you want to see all of the methods, including the one below, you can learn more about them in the documentation.
# Read lists of locations
with open('project_files/brent.txt', 'r') as f:
brent = f.read().splitlines()
with open('project_files/camden.txt', 'r') as f:
camden = f.read().splitlines()
with open('project_files/redbridge.txt', 'r') as f:
redbridge = f.read().splitlines()
with open('project_files/southwark.txt', 'r') as f:
southwark = f.read().splitlines()
Note that when text files are read using the splitlines() function, the resulting object is a list.
So the four objects you just created from the files - brent, camden, redbridge, and southwark - are all lists.
print( type(brent) )
print( type(camden) )
print( type(redbridge) )
print( type(southwark) )
<class 'list'> <class 'list'> <class 'list'> <class 'list'>
Print the number of locations in each listed borough:
# Print length of each list
print(len(brent))
print(len(camden))
print(len(redbridge))
print(len(southwark))
19 22 18 15
# Print the first 5 locations of each Borough
print(brent[:5])
print(camden[:5])
print(redbridge[:5])
print(southwark[:5])
['Brent Park', 'Church End', 'Dollis Hill', 'Harlesden', 'Kensal Green'] ['Chalk Farm', 'Dartmouth Park', 'Fitzrovia', 'Frognal', 'Bloomsbury'] ['Chadwell Heath', 'Gants Hill', 'Goodmayes', 'Snaresbrook', 'Hainault'] ['Denmark Hill', 'Dulwich', 'Surrey Quays', 'East Dulwich', 'Elephant and Castle']
# Is 'Newbury Park' in Redbridge?
print( 'Newbury Park' in redbridge ) # Membership
# Is 'Peckham' in Brent?
print( 'Peckham' in brent ) # Membership
True False
# Print minimum value in southwark
print( min(southwark)) # Min in this case is alphabet-sorted
# Print maximum value in southwark
print( max(southwark))
Bankside Walworth
Sets¶
Sets are unordered collections of unique objects, enclosed by curly braces: { }
For example:
integer_set = {0, 1, 2, 3, 4}
print( integer_set )
print( type(integer_set) )
{0, 1, 2, 3, 4}
<class 'set'>
Removing duplicates¶
Because each element in a set must be unique, sets are a great tool for removing duplicates. For example:
fibonacci_list = [ 1, 1, 2, 3, 5, 8, 13 ] # Will keep both 1's will remain
fibonacci_set = { 1, 1, 2, 3, 5, 8, 13 } # Only one 1 will remain
print( fibonacci_list )
print( fibonacci_set )
[1, 1, 2, 3, 5, 8, 13]
{1, 2, 3, 5, 8, 13}
Converting to sets¶
# Create a list
fibonacci_list = [ 1, 1, 2, 3, 5, 8, 13 ]
# Convert it to a set
fibonacci_set = set(fibonacci_list)
print( fibonacci_set )
{1, 2, 3, 5, 8, 13}
Set Operations¶
powers_of_two = { 1, 2, 4, 8, 16 }
fibonacci_set = { 1, 1, 2, 3, 5, 8, 13 }
# Union: Elements in either set (two ways, both do same thing)
print( powers_of_two.union( fibonacci_set ) )
print( powers_of_two | fibonacci_set )
{1, 2, 3, 4, 5, 8, 13, 16}
{1, 2, 3, 4, 5, 8, 13, 16}
# Intersection: Elements in both sets (two ways, both do same thing)
print( powers_of_two.intersection( fibonacci_set ) )
print( powers_of_two & fibonacci_set )
{8, 1, 2}
{8, 1, 2}
# Difference
print( powers_of_two )
print( fibonacci_set )
print( powers_of_two - fibonacci_set )
print( fibonacci_set - powers_of_two)
{1, 2, 4, 8, 16}
{1, 2, 3, 5, 8, 13}
{16, 4}
{13, 3, 5}
Analysis: remomving duplicates in our London data¶
Let's continue 007's training. Before we continue, lets see if wwe have duplicates and if so, remove them from our lists:
# Does Brent have duplicates?
print(len(brent) != len(set(brent)))
# Does Camden have duplicates?
print(len(camden) != len(set(camden)))
# Does Redbridge have duplicates?
print(len(redbridge) != len(set(redbridge)))
# Does Southwark have duplicates?
print(len(southwark) != len(set(southwark)))
False True True True
For the lists with duplicates, remove duplicates by converting them into sets. Then, convert them back into lists.
# Convert lists to sets to remove duplicates, then convert them back to lists
brent = list(set(brent))
camden = list(set(camden))
redbridge = list(set(redbridge))
southwark = list(set(southwark))
print(len(brent))
print(len(camden))
print(len(redbridge))
print(len(southwark))
19 19 16 13
Dictionaries¶
Dictionaries are unordered collections of key-value pairs, enclosed by curly braces, like {}.
integer_dict = {
'zero' : 0,
'one' : 1,
'two' : 2,
'three' : 3,
'four' : 4
}
print( integer_dict )
print( type(integer_dict) )
{'zero': 0, 'one': 1, 'two': 2, 'three': 3, 'four': 4}
<class 'dict'>
Keys and values¶
A dictionary, also called a "dict", is like a miniature database for storing and organizing data.
Each element in a dict is actually a key-value pair, and it's called an item.
- Key: A key is like a name for the value. You should make them descriptive.
- Value: A value is some other Python object to store. They can be floats, lists, functions, and so on.
Here's an example with descriptive keys:
my_dict = {
'title' : "The Foundation Trillogy",
'author' : 'Issac Assimov',
100 : ['A number.', 'The answer to 60 + 40 = ?'] # Keys can be integers too!
}
Access values using their Keys:
# Print the value for the 'title' key
print( my_dict['title'] )
# Print the value for the 'author' key
print( my_dict['author'] )
The Foundation Trillogy Issac Assimov
Updating values
# Updating existing key-value pair
my_dict['author'] = 'Isaac Asimov'
# Print the value for the 'author' key
print( my_dict['author'] )
Isaac Asimov
# Append element to list
my_dict[100].append('Answer to the Ultimate Question of Life, the Universe, and Everything')
# Print value for the key 42
print( my_dict[100] )
['A number.', 'The answer to 60 + 40 = ?', 'Answer to the Ultimate Question of Life, the Universe, and Everything']
Creating new items
# Creating a new key-value pair
my_dict['year'] = 1951
# Print summary of the book.
print('{} was written by {} in {}.'.format(my_dict['title'], my_dict['author'], my_dict['year']) )
The Foundation Trillogy was written by Isaac Asimov in 1951.
Convenience functions
# Keys - get entire list
print( my_dict.keys() )
# Values - get all values
print( my_dict.values() )
dict_keys(['title', 'author', 100, 'year']) dict_values(['The Foundation Trillogy', 'Isaac Asimov', ['A number.', 'The answer to 60 + 40 = ?', 'Answer to the Ultimate Question of Life, the Universe, and Everything'], 1951])
# All items (list of tuples)
print( my_dict.items() )
dict_items([('title', 'The Foundation Trillogy'), ('author', 'Isaac Asimov'), (100, ['A number.', 'The answer to 60 + 40 = ?', 'Answer to the Ultimate Question of Life, the Universe, and Everything']), ('year', 1951)])
Analysis: creating dictionaries and aggregating borough data¶
Lets create a locations dictionary for the London boroughs:
# Create location_dict
location_dict = {
'Brent' : brent,
'Camden' : camden,
'Redbridge' : redbridge,
'Southwark' : southwark
}
Lets double check that the dictionary has the correct keys using a for loop. More on loops syntax in later section:
for borough in ['Brent', 'Camden', 'Redbridge', 'Southwark']:
print( borough in location_dict )
True True True True
Since the dictionary is correct, we do not need the original lists, so we can get rid of them:
brent, camden, redbridge, southwark = None, None, None, None
type(brent)
NoneType
nb: None is an object that denotes emptiness.
Now, we want to split our visit to London into two trips: one for Inner London and one for Outer London.
Lets add two new items to our dictionary:
- Key:
'Inner London'... Value: All locations in'Camden'and'Southwark'. - Key:
'Outer London'... Value: All locations in'Brent'and'Redbridge'.
# Create a new key-value pair for 'Inner London'
location_dict['Inner London'] = location_dict['Camden'] + location_dict['Southwark']
# Create a new key-value pair for 'Outer London'
location_dict['Outer London'] = location_dict['Brent'] + location_dict['Redbridge']
# Quick QA - Add the locations of boroughs
len(location_dict['Camden']) + len(location_dict['Southwark']) + len(location_dict['Brent']) + len(location_dict['Redbridge'])
67
# Quick QA - Add innner and outer loactions
len(location_dict['Inner London']) + len(location_dict['Outer London'])
67
Save work using Pickle
- Use a Python built-in package called
pickleto save thelocation_dictobject to use later. - Pickle saves an entire object in a file on your computer.
- Pickling is a way to convert a python object (list, dict, etc.) into a character stream. The idea is that this character stream contains all the information necessary to reconstruct the object in another python script.
#Import pickle module
import pickle
# Save object to disk
with open('project_files/location_dict.pkl', 'wb') as f: # Saving the file 'location_dict' to project_files folder
pickle.dump(location_dict, f)
# Flow and Functions
if statements allow conditional logic¶
if...¶
If statements check if a condition is met before running a block of code.
Begin them with the if keyword. Then the statement has two parts:
- The condition, which must evaluate to a boolean. (Technically, it's fine as long as its "truthiness" can be evaluated.
- The code block to run if the condition is met (indented with 4 spaces).
For example:
current_fuel = 85
# Condition
if current_fuel >= 80:
# Code block to run if condition is met
print( 'We have enough fuel to last the zombie apocalypse. ')
We have enough fuel to last the zombie apocalypse.
if... else...¶
current_fuel = 50
# Condition
if current_fuel >= 80:
# Do this when condition is met
print( 'We have enough fuel to last the zombie apocalypse. ')
else:
# Do this when condition is not met
print( 'Restock! We need at least {} gallons.'.format(80 - current_fuel) )
Restock! We need at least 30 gallons.
if... elif... else...¶
current_fuel = 70
# First condition
if current_fuel >= 80:
print( 'We have enough fuel to last the zombie apocalypse. ')
# If first condition is not met, check this condition
elif current_fuel < 60:
print( 'ALERT: WE ARE WAY TOO LOW ON FUEL!' )
# If no conditions were met, perform this
else:
print( 'Restock! We need at least {} gallons.'.format(80 - current_fuel) )
Restock! We need at least 10 gallons.
Analysis: importing data and simple flow¶
We have created our location dictionary containing the boroughs we are interested in visiting. Now, we want to determine which location to visit first.
First, let's import location_dict again using pickle.
# Import location_dict using pickle
import pickle
# Read object from disk
with open('project_files/location_dict.pkl', 'rb') as f:
location_dict = pickle.load(f)
# Print the keys in location_dict
print(location_dict.keys())
dict_keys(['Brent', 'Camden', 'Redbridge', 'Southwark', 'Inner London', 'Outer London'])
Pseudocode for code below:
- If our Inner London list has more locations than our Outer London list, print the message:
Inner London is huge!
- Else if our Outer London list has more locations than our Inner London list, print the message:
Outer London is huge!
- Else (i.e. they have the same number of locations), print the message:
Inner and outer London are huge!
if len(location_dict['Inner London']) > len(location_dict['Outer London']):
print('Inner London is huge!')
elif len(location_dict['Outer London']) > len(location_dict['Inner London']):
print('Outer London is huge!')
else:
print('Inner and outer London are huge!')
Outer London is huge!
for loops allow iteration¶
for...¶
# Print list
for number in [0, 1, 2, 3, 4]:
print( number )
0 1 2 3 4
Range¶
# Print range
for number in range(5):
print( number )
0 1 2 3 4
# Print reversed range
for number in reversed(range(5)):
print( number )
4 3 2 1 0
Nested control flow¶
# Check if number divisible by another number
range_list = range(10)
for number in reversed(range_list):
if number == 0:
print( 'Liftoff!' )
elif number % 3 == 0:
print( 'Buzz' )
else:
print( number )
Buzz 8 7 Buzz 5 4 Buzz 2 1 Liftoff!
# for loops within other for loops
list_a = [4, 3, 2]
list_b = [6, 3]
for a in list_a:
for b in list_b:
print( a, 'x', b, '=', a * b )
4 x 6 = 24 4 x 3 = 12 3 x 6 = 18 3 x 3 = 9 2 x 6 = 12 2 x 3 = 6
Building new lists¶
- Set a variable to an empty list, which is just
[]. - Then, as you loop through elements,
.append()the ones you want to your list variable.
# Separate range_list into even_list and odd_list
range_list = range(10)
# Initialize empty lists
evens_list = []
odds_list = []
# Iterate through each number in range_list
for number in range_list:
# check for divisibility by 2
if number % 2 == 0:
# If divisible by 2
evens_list.append(number)
else:
# If not divisible by 2
odds_list.append(number)
# Confirm our lists are correct
print( evens_list )
print( odds_list )
[0, 2, 4, 6, 8] [1, 3, 5, 7, 9]
Analysis: Counting Locations in London¶
Lets count the number of locations in each of our boroughs.
- Tip: You can iterate through keys and values of a dictionary at the same time using
.items(), like so:
for key, value in location_dict.items():
# code block
- Tip: Remember, to insert multiple dynamic values into a string, you can just add more places to
.format(), like so:
'{} has {} locations.'.format(first_value, second_value)
# For each key in location_dict, print the number of locations in its list
for key, value in location_dict.items():
print('{} has {} locations.'.format(key, len(value)))
Brent has 19 locations. Camden has 19 locations. Redbridge has 16 locations. Southwark has 13 locations. Inner London has 32 locations. Outer London has 35 locations.
Now, let's give each location in Outer London a first impression based on its name.
Pseudocode
Combine if and for statements. For each location in Outer London...
- If its name has
'Farm','Park','Hill', or'Green'in it, print:
{name} sounds pleasant.
- Else If its name has
'Royal','Queen', or'King'in it, print:
{name} sounds grand.
- If its name doesn't sound pleasant or grand, just ignore it.
- Tip: If you want to check if any word from a list is found in a string, you can use
any(), like so:
any( word in name for word in list_of_words )
pleasant_sounding = ['Farm', 'Park', 'Hill', 'Green']
royal_sounding = ['Royal', 'Queen', 'King']
# Print first impression of each location in Outer London based on its names
for name in location_dict['Outer London']:
if any(word in name for word in pleasant_sounding): # 'word' is part of any function
print( name, 'sounds pleasant.' )
elif any(word in name for word in royal_sounding):
print( name, 'sounds grand.' )
Kensal Green sounds pleasant. Wembley Park sounds pleasant. Queen's Park sounds pleasant. Dollis Hill sounds pleasant. Brent Park sounds pleasant. Park Royal sounds pleasant. Kingsbury sounds grand. Woodford Green sounds pleasant. Seven Kings sounds grand. Newbury Park sounds pleasant. Gants Hill sounds pleasant.
List comprehensions construct new lists elegantly¶
List comprehensions¶
List comprehensions aren't mandatory, but they help keep your code clean and concise.
List comprehensions construct new lists out of existing ones after applying transformations or conditions to them.
Example
# Construct list of the squares in range(10) using list comprehension
squares_list = [number**2 for number in range(10)]
print( squares_list )
[0, 1, 4, 9, 16, 25, 36, 49, 64, 81]
Conditional inclusion¶
You can even include conditional logic in list comprehensions!
# Conditional inclusion
evens_list = [number for number in range(10) if number % 2 == 0]
print( evens_list )
[0, 2, 4, 6, 8]
Conditional outputs¶
# Conditional outputs
even_odd_labels = ['Even' if number % 2 == 0 else 'Odd' for number in range(10)]
print( even_odd_labels )
['Even', 'Odd', 'Even', 'Odd', 'Even', 'Odd', 'Even', 'Odd', 'Even', 'Odd']
Other comprehensions¶
Finally, list comprehensions are not limited to lists!
- You can use them for other data structures too.
- The syntax is the same, except you would enclose it with curly braces for sets and parentheses for tuples.
For example, we can create a set like so:
# Construct set of doubles using set comprehension
doubles_set = { number * 2 for number in range(10) }
print( doubles_set )
{0, 2, 4, 6, 8, 10, 12, 14, 16, 18}
Analysis: list comprehensions of London locations¶
Lets use a list comprehension, to create a new list called pleasant_locations.
- It should contain locations in Outer London that are
pleasant_sounding.
# Create pleasant_locations list using a list comprehension
pleasant_locations = [name for name in location_dict['Outer London']
if any(word in name for word in pleasant_sounding)]
# Print the pleasant-sounding locations
print(pleasant_locations)
['Kensal Green', 'Wembley Park', "Queen's Park", 'Dollis Hill', 'Brent Park', 'Park Royal', 'Woodford Green', 'Newbury Park', 'Gants Hill']
# Print number of pleasant-sounding locations
len(pleasant_locations)
9
Functions are blocks of reusable code¶
def make_message_exciting(message='hello, world'):
text = message + '!'
return text
print( type(make_message_exciting) )
<class 'function'>
Let's break that down:
- Functions begin with the
defkeyword, followed by the function name - They can take optional arguments, E.g., message = 'hello world'
- They are then followed by an indented code block.
- Finally, they return a value, which is also indented
To call a function, simply type its name and a parentheses.
# Call make_message_exciting() function
make_message_exciting()
'hello, world!'
In practice¶
In practice, functions are ideal for isolating functionality.
def square(x):
output = x*x
return output
def cube(x):
output = x*x*x
return output
print( square(3) )
print( cube(2) )
print( square(3) + cube(2) )
9 8 17
Optional parts¶
nb: code block in a function is actually optional, as long as you have a return statement.
# Example of function without a code block
def hello_world():
return 'hello world'
# Call hello_world() function
hello_world()
'hello world'
And also... the return statement is optional, as long as you have a code block.
- If no return statement is given, the function will return
Noneby default. - Code blocks in the function will still run.
# Example of function without a return statement
def print_hello_world():
print( 'hello world' )
# Call print_hello_world() function
print_hello_world()
hello world
Arguments¶
Arguments can have also default values, set using the = operator.
def print_message( message='Hello, world', punctuation='.' ):
output = message + punctuation
print( output )
# Print default message
print_message()
Hello, world.
To pass a new value for the argument, simply set it again when calling the function.
# Print new message, but default punctuation
print_message( message='Nice to meet you', punctuation='...' )
Nice to meet you...
When passing a value to an argument, you don't have to write the argument's name if the values are in order.
# Print new message without explicity setting the argument
print_message( 'Where is everybody', '?' )
Where is everybody?
Analysis: Using functions¶
Lets write a function called filter_locations that takes two arguments:
location_listwords_list
The function should return the list of names in location_list that have any word in words_list.
def filter_locations(location_list, words_list):
return [name for name in location_list
if any(word in name for word in words_list)]
Next, let's test that function.
**Lets create a new
pleasant_locations list using the function we just wrote.**
* Pass in the list of Outer London locations and the list of pleasant-sounding words.
# Create pleasant_locations using filter_locations()
pleasant_locations = filter_locations(location_dict['Outer London'], pleasant_sounding)
# Print list of pleasant-sounding locations
print( pleasant_locations )
['Kensal Green', 'Wembley Park', "Queen's Park", 'Dollis Hill', 'Brent Park', 'Park Royal', 'Woodford Green', 'Newbury Park', 'Gants Hill']
Next, let's use this handy function to create a grand_locations list for locations that sound grand.
- Pass in the list of Outer London locations and the list of grand-sounding words.
# Create grand_locations using filter_locations()
grand_locations = filter_locations(location_dict['Outer London'], royal_sounding)
# Print list of grand-sounding locations
print(grand_locations)
["Queen's Park", 'Park Royal', 'Kingsbury', 'Seven Kings']
Awesome, are there any locations in both lists?
**Display the locations in both the
pleasant_locations and grand_locations lists.**
* You can compare these lists manually because they are short, but try using sets and the .intersection() function (or & operator)!
# Display locations that sound pleasant and grand (two ways)
# Using .intersection() function
print(set(pleasant_locations).intersection( set(grand_locations) ))
# Using & operator
print(set(pleasant_locations) & set(grand_locations)) # I prefer this!
{"Queen's Park", 'Park Royal'}
{"Queen's Park", 'Park Royal'}
Great, we'll start with these for our visit.
# NumPy
Let's import NumPy.
import numpy as np # In code to left, we set an alias for NumPy to np
Arrays are homogeneous¶
NumPy Arrays¶
NumPy arrays are tables of elements that all share the same data type (usually numeric).
numpy.ndarray is its official type.
# Array of ints
array_a = np.array([0, 1, 2, 3])
print( array_a )
print( type(array_a) )
[0 1 2 3] <class 'numpy.ndarray'>
# Print data type of contained elements
print( array_a.dtype )
int64
- The NumPy array array_a itself has type numpy.ndarray.
- The elements contained inside the array have type int64.
Homogenous¶
NumPy arrays are homogenous, which means all of their elements must have the same data type.
Because NumPy doesn't support mixed types, it converts all of the elements in a mixed array into strings.
See following:
# Mixed array with 1 string and 2 integers
array_b = np.array(['four', 5, 6])
# Print elements in array_b
print( array_b )
['four' '5' '6']
Shape¶
The two arrays we created above, array_a and array_b both have only 1 axis.
- You can think an axis as a direction in the coordinate plane.
- For example, lines have 1 axis, squares have 2 axes, cubes have 3 axes, etc.
We can use the .shape attribute to see the axes for a NumPy array.
print( array_a.shape )
print( array_b.shape )
(4,) (3,)
As you can see, .shape returns a tuple.
- The number of elements in the tuple is the number of axes.
- And each element's value is the length of that axis.
Together, these two pieces of information make up the shape, or dimensions, of the array.
- So array_a is a 4x1 array. It has 1 axis of length 4.
- And array_b is a 3x1 array. It has 1 axis of length 3.
- They are both considered "1-dimensional" arrays.
Indexing¶
print(array_a)
# First element of array_a
print( array_a[0] )
# Last element of array_a
print( array_a[-1] )
[0 1 2 3] 0 3
# Or can SLICE it: From second element of array_a up to the 4th
print( array_a[2:4] )
[2 3]
Missing data¶
- NumPy has a special
np.nanobject for denoting missing values. - This object is called
NaN, which stands for Not a Number.
For example, let's create an array with a missing value:
# Array with missing values
array_with_missing_value = np.array([1.2, 8.8, 4.0, np.nan, 6.1])
# Print array
print( array_with_missing_value )
[ 1.2 8.8 4. nan 6.1]
# Print array's dtype
print( array_with_missing_value.dtype )
float64
Analysis: Building indices locations database¶
*In the previous analysis...*
In the previous analysis, we decided on Park Royal as our first stop in our training mission to London.
Now we've arrived at the tube station!

Park Royal Tube Station
Park Royal is home to London's largest business park.
- The business park supports around 1,700 businesses.
- Q hands you a manual with their names and locations.
- They are listed in order from smallest to largest (this will be important later).
**First, let's create a NumPy array called
business_ids.**
* The first business has ID 1, the second one has ID 2 and so on.
# Create array of business_ids with values ranging from 1 to 1700
business_ids = np.array(range(1, 1701))
Next, print the shape of business_ids to confirm it has shape (1700, ).
# Print shape of business_ids
print( business_ids.shape )
(1700,)
Finally, print the last 10 business ID's to confirm the array is set up properly.
# Print last 10 business ID's
print( business_ids[-10:] )
# Wrong wway to do it
print( business_ids[-10:-1] ) # Remember, the end number not included!
[1691 1692 1693 1694 1695 1696 1697 1698 1699 1700] [1691 1692 1693 1694 1695 1696 1697 1698 1699]
Arrays are multidimensional¶
Multidimensional array¶
For creating matrices!
For example, let's create a matrix with 2 rows and 3 columns:
array_c = np.array([[1, 2, 3], [4, 5, 6]])
print( array_c )
[[1 2 3] [4 5 6]]
# The NumPy array above is 2x3.
print( array_c.shape )
(2, 3)
Reshape¶
For example, we can reshape it from its original shape of 2x3 to the new shape of 3x2:
# Reshape to 3x2
print( array_c.reshape(3, 2) )
[[1 2] [3 4] [5 6]]
Nb: this is not the same as transposing array_c, because we are keeping the same order of the elements.
# Reshape to 1x6
print( array_c.reshape(1,6) )
[[1 2 3 4 5 6]]
# Print number axes
print(array_c)
print( len(array_c.reshape(1,6).shape) )
[[1 2 3] [4 5 6]] 2
Flatten¶
If you want to reshape it to 1x6 and reduce the number of axes, then you actually need to reshape it to (6,) with an empty second axis.
- There's a shortcut for reducing an array to 1 axis: the
.flatten()function.
# Reshape and reduce axes
print( array_c.reshape(6,) )
# Flatten (reduce to 1 axis) # Shortcut
print( array_c.flatten() )
# Print number of axes
print( len(array_c.reshape(6,).shape) )
print( len(array_c.flatten().shape) )
[1 2 3 4 5 6] [1 2 3 4 5 6] 1 1
# Nb: the original function is immutable!
print(array_c)
print(array_c.shape)
[[1 2 3] [4 5 6]] (2, 3)
2.4 - Transpose¶
If you do want to transpose the array, you can simply .transpose() it!
- Transposing will flip the rows and columns of an array (not just the shape).
- Reshaping will simply change the shape, but keep the same order of elements.
# Transpose
print( array_c.transpose() )
# REMEMBER the original data is still the same
print(array_c)
[[1 4] [2 5] [3 6]] [[1 2 3] [4 5 6]]
Indexing¶
Let's look at an example with a 3x3 array:
# Create a 3x3 array
array_d = np.array(range(1,10)).reshape(3,3)
print(array_d)
[[1 2 3] [4 5 6] [7 8 9]]
# Select all elements in the first row
# Tip: A colon (:) means select all elements along that axis.
print( array_d[0, :] )
[1 2 3]
# Select second row and all columns
print( array_d[1, :] )
[4 5 6]
# Select third column and all rows
print( array_d[:, 2])
[3 6 9]
# Select second column and all rows
print( array_d[:, 1])
[2 5 8]
# Select after second row and second column
print( array_d[1:, 1:] )
[[5 6] [8 9]]
Analysis: Sampling¶
There are 1700 businesses in Park Royal's business park. However, you only have 10 hours to stay in Park Royal before you need to move on to the next location. Assuming you could only visit 5 businesses per hour, which businesses should you visit?
Well, 2 options immediately come to mind:
- You could just visit the first 50 businesses.
- You could randomly sample 50 businesses.
While these appear fine at first glance, there are potential flaws with both of these approaches. Remember, the businesses were listed in order from smallest to largest! Therefore...
- Just visiting the first 50 would give us a biased sample of only the smallest businesses.
- Visiting a random sample of 50 is better, but there's still a chance that our sample ends up biased from pure chance.
Instead, let's take a stratified random sample based on the size of the business.
WTF is stratified random sampling?¶
This is a very important concept for practical machine learning.
> *Stratified random sampling is first grouping your observations based on a key variable, and then randomly sampling from those groups.*
This will ensure your sample is representative of the broader dataset along that key variable.
Stratified Random Sampling
For this analysis, it just means that...
- We'll start by splitting our businesses into 10 groups of 170 businesses each.
- The first group will have ID's from 1 to 170, the second will have ID's from 171 to 340, etc.
- Then we'll randomly select 5 businesses from each group of 170.
- Since the businesses are already ordered by size, this will ensure that small, medium, and big businesses are all represented in our sample.
**First, reshape the 1-dimensional
business_ids array into a new 2-dimensional id_matrix array.**
# Create id_matrix by reshaping business_ids to have 10 columns
id_matrix = business_ids.reshape(170,10)
# Print shape
print(id_matrix.shape)
(170, 10)
print(id_matrix)
[[ 1 2 3 ..., 8 9 10] [ 11 12 13 ..., 18 19 20] [ 21 22 23 ..., 28 29 30] ..., [1671 1672 1673 ..., 1678 1679 1680] [1681 1682 1683 ..., 1688 1689 1690] [1691 1692 1693 ..., 1698 1699 1700]]
Great, now we have a matrix with 10 columns representing 10 groups of businesses. But remember, our goal is to stratify our sample by size of the business, and because our businesses are ordered by size, the first group should be 1 to 170, the second group should be 171 to 340, and so on.
**Print the first column (group) of
id_matrix.**
* Does it contain businesses 1 to 170?
# Print first column of id_matrix
print(id_matrix[:,0])
[ 1 11 21 31 41 51 61 71 81 91 101 111 121 131 141 151 161 171 181 191 201 211 221 231 241 251 261 271 281 291 301 311 321 331 341 351 361 371 381 391 401 411 421 431 441 451 461 471 481 491 501 511 521 531 541 551 561 571 581 591 601 611 621 631 641 651 661 671 681 691 701 711 721 731 741 751 761 771 781 791 801 811 821 831 841 851 861 871 881 891 901 911 921 931 941 951 961 971 981 991 1001 1011 1021 1031 1041 1051 1061 1071 1081 1091 1101 1111 1121 1131 1141 1151 1161 1171 1181 1191 1201 1211 1221 1231 1241 1251 1261 1271 1281 1291 1301 1311 1321 1331 1341 1351 1361 1371 1381 1391 1401 1411 1421 1431 1441 1451 1461 1471 1481 1491 1501 1511 1521 1531 1541 1551 1561 1571 1581 1591 1601 1611 1621 1631 1641 1651 1661 1671 1681 1691]
Crap, that's not what we wanted.
Remember, when you .reshape an array, the new array keeps the order of the elements.
Interlude: Toy Problem
Let's walk through a miniature example of what we just did in previous analysis, because it will be easier to understand by peeking under the hood.
- Instead of 1700 businesses, let's say we only had 170 businesses to visit, but we still want to group them into 10 groups by ID.
- Therefore, we want the first group to have businesses 1 to 17, the second group to have 18 to 34, the third group to have 35 to 51, and so on.
# Create array of 170 ID's
mini_business_ids = np.array( range(1, 171) )
# Reshape to have 10 columns
mini_id_matrix = mini_business_ids.reshape(17, 10)
# Display mini_id_matrix
print( mini_id_matrix )
[[ 1 2 3 4 5 6 7 8 9 10] [ 11 12 13 14 15 16 17 18 19 20] [ 21 22 23 24 25 26 27 28 29 30] [ 31 32 33 34 35 36 37 38 39 40] [ 41 42 43 44 45 46 47 48 49 50] [ 51 52 53 54 55 56 57 58 59 60] [ 61 62 63 64 65 66 67 68 69 70] [ 71 72 73 74 75 76 77 78 79 80] [ 81 82 83 84 85 86 87 88 89 90] [ 91 92 93 94 95 96 97 98 99 100] [101 102 103 104 105 106 107 108 109 110] [111 112 113 114 115 116 117 118 119 120] [121 122 123 124 125 126 127 128 129 130] [131 132 133 134 135 136 137 138 139 140] [141 142 143 144 145 146 147 148 149 150] [151 152 153 154 155 156 157 158 159 160] [161 162 163 164 165 166 167 168 169 170]]
Aha, now we're on to something... so instead of reshaping to 17 rows and 10 columns, what if we reshape to 10 rows and 17 columns?
# Reshape to have 10 rows
mini_id_matrix = mini_business_ids.reshape(10, 17)
# Display mini id matrix
print( mini_id_matrix )
[[ 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17] [ 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34] [ 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51] [ 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68] [ 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85] [ 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102] [103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119] [120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136] [137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153] [154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170]]
Now we have 10 rows instead of 10 columns.
- The first row has the businesses 1 to 17, which is what we want!
- And if we want the first column to have businesses 1 to 17, we can simply transpose this matrix.
Nb: Toy problems are miniature versions of your problem that are easier to break apart and understand conceptually.
Let's try building that id_matrix again.
**This time, reshape it to have 10 rows, each representing 1 group of businesses.** * The first row should contain businesses 1 to 170.
# Reshape business_ids to have 10 rows, with 170 businesses each
id_matrix = business_ids.reshape(10,170)
print(id_matrix)
[[ 1 2 3 ..., 168 169 170] [ 171 172 173 ..., 338 339 340] [ 341 342 343 ..., 508 509 510] ..., [1191 1192 1193 ..., 1358 1359 1360] [1361 1362 1363 ..., 1528 1529 1530] [1531 1532 1533 ..., 1698 1699 1700]]
# Print the first row of id_matrix
print(id_matrix[0,:])
[ 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170]
We now have businesses with ID's from 1 to 170.
Now, what if we still want our groups to be grouped by column, instead of by row? Let's flip our rows and columns now that the rows have the correct groups.
**Overwrite
id_matrix with its transposed version.**
# Overwrite id_matrix with flipped version
id_matrix = id_matrix.transpose()
# Print first column
print(id_matrix)
# Print shape of new id_matrix
print(id_matrix.shape)
[[ 1 171 341 ..., 1191 1361 1531] [ 2 172 342 ..., 1192 1362 1532] [ 3 173 343 ..., 1193 1363 1533] ..., [ 168 338 508 ..., 1358 1528 1698] [ 169 339 509 ..., 1359 1529 1699] [ 170 340 510 ..., 1360 1530 1700]] (170, 10)
Math is elementwise¶
- Functions are applied to each element in the array.
- Operations are applied between corresponding elements in two arrays.
Example:
# 2x2 matrix of floats
x = np.array([[1.0, 2.0], [3.0, 4.0]])
print( x )
# 2x2 matrix of floats
y = np.array([[2.0, 5.0], [10.0, 3.0]])
print( y )
[[ 1. 2.] [ 3. 4.]] [[ 2. 5.] [ 10. 3.]]
Between two arrays¶
# Addition
print( x + y ) # This is matrices addition, from linear algebra!
[[ 3. 7.] [ 13. 7.]]
# Subtraction
print( x - y )
[[-1. -3.] [-7. 1.]]
# Multiplication
print( x * y )
[[ 2. 10.] [ 30. 12.]]
# Division
print( x / y )
[[ 0.5 0.4 ] [ 0.3 1.33333333]]
# Modulo
print( x % y )
[[ 1. 2.] [ 3. 1.]]
Between array and scalar¶
print(x)
[[ 1. 2.] [ 3. 4.]]
# Addition
print( x + 2 )
[[ 3. 4.] [ 5. 6.]]
# Subtraction
print( x - 2 )
[[-1. 0.] [ 1. 2.]]
# Multiplication
print( x * 2 )
[[ 2. 4.] [ 6. 8.]]
# Modulo
print( x % 2 )
[[ 1. 0.] [ 1. 0.]]
Math functions¶
A few of the most useful ones include:
np.sqrt()for square rootnp.abs()for absolute valuenp.power()for raising elements to a powernp.exp()for calculating exponentialsnp.log()for calculating the natural log
Example
# Cubed
print(x)
np.power(x, 3)
[[ 1. 2.] [ 3. 4.]]
array([[ 1., 8.],
[ 27., 64.]])
Aggregation functions¶
A few of the most useful ones include:
np.sum()for calculating the sumnp.min()for finding the smallest elementnp.max()for finding the largest elementnp.median()for finding the mediannp.mean()for calculating the meannp.std()for calculating the standard deviation
Example:
print( x )
# Sum of all elements
print( np.sum(x) )
[[ 1. 2.] [ 3. 4.]] 10.0
Axis¶
Now, what if we want to aggregate across columns or rows, instead of across the entire array?
We can pass in an axis argument:
axis=0to aggregate across columnsaxis=1to aggregate across rows
For example:
print( x )
# Sum of each column
print( np.sum(x, axis=0) )
[[ 1. 2.] [ 3. 4.]] [ 4. 6.]
# Sum of each row
print( np.sum(x, axis=1) )
[ 3. 7.]
Analysis: finding min and max¶
Let's take another look at our id_matrix.
id_matrix
array([[ 1, 171, 341, ..., 1191, 1361, 1531],
[ 2, 172, 342, ..., 1192, 1362, 1532],
[ 3, 173, 343, ..., 1193, 1363, 1533],
...,
[ 168, 338, 508, ..., 1358, 1528, 1698],
[ 169, 339, 509, ..., 1359, 1529, 1699],
[ 170, 340, 510, ..., 1360, 1530, 1700]])
- In previous analysis we displayed the first group (column) and confirmed that the ID's were from 1 to 170.
- Now let's confirm the rest of the array is correct by finding the
np.min()andnp.max()of each group.
**First, create and print an object called
group_min with the minimum ID of each group (column) of id_matrix.**
# Create group_min of each column
group_min = np.min(id_matrix, axis=0)
# Print group_min
print(group_min)
[ 1 171 341 511 681 851 1021 1191 1361 1531]
Next, lets create and print an object called group_max with the maximum ID of each group (column) of id_matrix.
# Create group_max of each column
group_max = np.max(id_matrix, axis=0)
# Print group_max
print(group_max)
[ 170 340 510 680 850 1020 1190 1360 1530 1700]
Finally, subtract group_min from group_max to confirm that each of the 10 groups has a range of 170.
- Remember to add 1 to the difference between max and min because the ends are inclusive
- i.e. 170 - 1 = 169
# Print range of each group
print(group_max - group_min +1)
[170 170 170 170 170 170 170 170 170 170]
NumPy is reliably random¶
NumPy comes with a submodule called np.random.
- This submodule has many useful tools for random sampling from various distributions and generating randomized data.
Interlude: Tab trick
Pressing tab after typeing np.random., will show all the different methods! This is called tab completion.
Random array¶
Let's try generating a 8x8 matrix of random digits from 0 to 9.
We can use the np.random.randomint() function, which draws from a uniform distribution (i.e. each integer is equally likely to be drawn).
It takes 3 arguments:
- low: Lower boundary for sampling (inclusive)
- high: Upper boundary for sampling (exclusive)
- size: Number of samples to draw
First, let's generate 64 random digits:
# Randomly draw 64 samples from a uniform distribution from [0, 10]
sample = np.random.randint(low=0, high=10, size=64)
print( sample )
[1 8 8 3 0 1 3 5 4 3 9 2 2 8 0 3 4 3 7 9 0 5 8 2 7 4 6 5 4 4 3 7 2 8 2 1 8 4 8 2 6 7 9 0 0 2 3 5 9 6 1 5 5 5 3 0 6 5 8 8 6 6 4 8]
Next, we can reshape that array into the 8x8 matrix we wanted.
print(sample.reshape(8,8))
[[1 8 8 3 0 1 3 5] [4 3 9 2 2 8 0 3] [4 3 7 9 0 5 8 2] [7 4 6 5 4 4 3 7] [2 8 2 1 8 4 8 2] [6 7 9 0 0 2 3 5] [9 6 1 5 5 5 3 0] [6 5 8 8 6 6 4 8]]
Random multidimensional array¶
However, there's an easier way to create a 8x8 matrix right from the start.
The size argument of np.random.randomint() can actually accept a tuple with one value per axis, like so:
# Randomly draw 8x8 samples from a uniform distribution from [0, 10]
generated_matrix = np.random.randint(0, 10, (8,8))
print( generated_matrix )
[[3 2 8 2 4 1 0 8] [0 5 1 8 6 4 3 0] [8 8 7 9 7 8 7 6] [4 0 5 9 4 7 1 2] [6 8 4 9 4 5 2 6] [4 2 3 2 5 2 9 1] [7 6 2 5 9 4 2 9] [4 6 3 5 1 4 9 1]]
nb: every time you run the code above, will generate new sample values!
Random seed¶
In machine learning, you'll often want to be able to reproduce your random samples. For example, if you need to close your file and come back to it later, you'll want to draw the same random samples so you can get consistent results.
That's where np.random.seed() comes in.
# Set seed for reproducible results
np.random.seed(1337)
# Randomly draw 8x8 samples from a uniform distribution from [0, 10)
generated_matrix = np.random.randint(0, 10, (8,8))
print( generated_matrix )
[[7 8 7 9 7 2 2 4] [8 9 6 6 7 8 1 6] [6 2 2 9 8 1 7 3] [1 3 9 3 4 8 8 7] [9 4 1 6 4 4 2 5] [7 4 2 3 9 2 4 8] [6 3 8 8 7 4 3 2] [9 1 2 0 3 8 2 1]]
Random choice¶
Next, what if we want to randomly select 5 elements from the first column of generated_matrix?
# Select first column of generated_matrix
print( generated_matrix[:,0] )
[7 8 6 1 9 7 6 9]
Well, if we want to randomly select 5 elements from that column, we can use np.random.choice()
# Set seed for reproducible results
np.random.seed(55)
# Randomly select 5 elements from first column of generated_matrix
print( np.random.choice(generated_matrix[:,0], 5) )
[7 6 9 7 7]
Replacement¶
By default, np.random.choice() samples with replacement.
- This just means that after each element is selected, it is replenished before selecting the next one.
- The third argument to
np.random.choice()tells it to sample with replacement or not.
Example (to ensure no elements are repeated):
# Set seed for reproducible results
np.random.seed(55)
# Randomly select 5 elements from first column of generated_matrix
print( np.random.choice(generated_matrix[:,0], 5, replace=False) )
[9 9 6 8 1]
Analysis: Random selection¶
We are now ready to select 5 businesses from each group in id_matrix.
Pseudocode for code below
Lets write a loop that chooses 5 businesses from column of id_matrix.
- Set the random seed to 9001.
- Sample without replacement.
# Seed random seed
np.random.seed(9001)
# Print selected businesses from each group
print( np.random.choice(id_matrix[:,0], 5, replace=False) )
[ 7 37 124 41 17]
# Seed random seed
np.random.seed(9001)
# Print selected businesses from each group
for group in range(id_matrix.shape[1]):
print('Group {}: {}'.format( group + 1, np.random.choice( id_matrix[:,group], 5, replace=False ) ) )
Group 1: [ 7 37 124 41 17] Group 2: [302 261 257 323 234] Group 3: [464 486 463 440 474] Group 4: [645 582 666 631 553] Group 5: [699 738 705 760 792] Group 6: [ 995 1000 909 869 899] Group 7: [1042 1058 1172 1122 1153] Group 8: [1304 1275 1343 1344 1236] Group 9: [1401 1417 1383 1387 1474] Group 10: [1642 1687 1545 1644 1549]
Awesome, now we're ready to start visiting the businesses... Let's start with Group 1 and the business with ID 7. Lets call this Casey's Flower Shop.
# Pandas
Let's fisrt import Pandas:
import pandas as pd
Pandas DataFrames¶
DataFrames are like highly optimized spreadsheets.
One of the most common ways to create a DataFrame is to pass a dictionary into the pd.DataFrame() function, like so:
example_dataframe = pd.DataFrame({
'column_1' : [5, 4, 3],
'column_2' : ['a', 'b', 'c']
})
example_dataframe
| column_1 | column_2 | |
|---|---|---|
| 0 | 5 | a |
| 1 | 4 | b |
| 2 | 3 | c |
Importing¶
- The one most useful way for us is by importing CSV (comma-separated) files.
- Pandas has a handy
pd.read_csv()function just for this purpose. - Pandas can also read from Excel, JSON, SQL, and many other formats.
Let's see an example:
# Read the iris dataset from a CSV file in our project_files folder
df = pd.read_csv('project_files/iris.csv')
# Print data type for df
print( type(df) )
<class 'pandas.core.frame.DataFrame'>
Head¶
# Display the first 5 rows of the dataframe
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
Shape¶
- Pandas DataFrames always have 2 axes (i.e. like a spreadsheet).
- The first value in the shape tuple is the number of rows, and the second is the number of columns.
# Shape of dataframe
print( df.shape )
(150, 5)
# Number of rows (both of below are equivalent)
print( len(df) )
print( df.shape[0] )
150 150
Summary statistics¶
# Print min value of each column in data frame df
print( df.min() )
sepal_length 4.3 sepal_width 2 petal_length 1 petal_width 0.1 species setosa dtype: object
# Print max value of each column in data frame df
print( df.max() )
sepal_length 7.9 sepal_width 4.4 petal_length 6.9 petal_width 2.5 species virginica dtype: object
Finally, one very quick way to summarize your data is with the .describe() function.
It will display many statistics at once, including:
- mean
- standard deviation
- mininum
- 25th percentile
- median
- 75th percentile
- maximum
of each column:
# Display summary statistics for each variable
df.describe()
| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| count | 150.000000 | 150.000000 | 150.000000 | 150.000000 |
| mean | 5.843333 | 3.057333 | 3.758000 | 1.199333 |
| std | 0.828066 | 0.435866 | 1.765298 | 0.762238 |
| min | 4.300000 | 2.000000 | 1.000000 | 0.100000 |
| 25% | 5.100000 | 2.800000 | 1.600000 | 0.300000 |
| 50% | 5.800000 | 3.000000 | 4.350000 | 1.300000 |
| 75% | 6.400000 | 3.300000 | 5.100000 | 1.800000 |
| max | 7.900000 | 4.400000 | 6.900000 | 2.500000 |
Analysis: Exploring the data¶
*In the previous analysis...*
- we discovered that Casey's Flower Shop is the first local business we need to visit in Park Royal.
When we arrive, we find out that the owner, Casey, urgently needs our help!
- They just received a new shipment of Iris flowers, but they've never stocked these flowers before.
- Casey asks us to share what we know about these new flowers.
- Luckily, we have the Iris dataset to learn more about them ourselves!
While 150 observations is not exactly "big data," it's still too large to fit on our screen at once. So let's use another toy problem to practice the concepts.
First, lets create a new DataFrame called
toy_df. It will contain the first 5 rows plus the last 5 rows from our original Iris dataset.
toy_df = pd.concat([df.head(), df.tail()]) #pd.concat combines or concatinates the top five and last five
Next, display toy_df
# Display toy_df
toy_df
| sepal_length | sepal_width | petal_length | petal_width | species | |
|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | virginica |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | virginica |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | virginica |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | virginica |
Next, display a summary table for toy_df.
- It will show the mean, standard deviation, and quartiles for each of the columns
# Describe toy_df
toy_df.describe()
| sepal_length | sepal_width | petal_length | petal_width | |
|---|---|---|---|---|
| count | 10.000000 | 10.000000 | 10.000000 | 10.000000 |
| mean | 5.590000 | 3.130000 | 3.290000 | 1.130000 |
| std | 0.807534 | 0.316403 | 1.995244 | 0.992248 |
| min | 4.600000 | 2.500000 | 1.300000 | 0.200000 |
| 25% | 4.925000 | 3.000000 | 1.400000 | 0.200000 |
| 50% | 5.500000 | 3.050000 | 3.250000 | 1.000000 |
| 75% | 6.275000 | 3.350000 | 5.175000 | 1.975000 |
| max | 6.700000 | 3.600000 | 5.400000 | 2.300000 |
Series are single columns¶
Pandas Series¶
- Series are single columns of data from DataFrames.
pandas.core.series.Seriesis the official type.- You can directly create a Series from a list. For example:
integer_series = pd.Series([0, 1, 2, 3, 4])
print( integer_series )
print( type(integer_series) )
0 0 1 1 2 2 3 3 4 4 dtype: int64 <class 'pandas.core.series.Series'>
Selecting from DataFrames¶
To select petal_length (two ways):
# Way 1
print( type(df.petal_length) )
# Way 2
print( type(df['petal_length']) )
# Check that both ways are identical
print( all(df.petal_length == df['petal_length']) ) #Setting to all returns True if all true, or False otherwise
<class 'pandas.core.series.Series'> <class 'pandas.core.series.Series'> True
Series functionality¶
# First 5 values of petal length
print( df.petal_length.head() )
# Minimum sepal length
print( 'The minimum petal length is', df.petal_length.min() )
# Maximum sepal length
print( 'The maximum petal length is', df.petal_length.max() )
0 1.4 1 1.4 2 1.3 3 1.5 4 1.4 Name: petal_length, dtype: float64 The minimum petal length is 1.0 The maximum petal length is 6.9
Unique values¶
For categorical (i.e. non-numeric) variables, it's useful to know which unique values they have.
- In the Iris dataset, the only categorical variable is 'species'
- Each unique value is also called a class.
- By the way, building a model to predict a categorical variable is called classification.
To find the unique classes, you can use the .unique() function:
# Print unique species
print( df.species.unique() )
['setosa' 'versicolor' 'virginica']
Series math¶
Series behave very similarly to 1-dimensional arrays from NumPy.
For example, you can directly apply many NumPy math functions to Pandas Series.
# Pandas
print( df.petal_length.mean() )
# NumPy
import numpy as np
print( np.mean( df.petal_length ) )
3.758 3.758
# Create new petal area feature
df['petal_area'] = df.petal_width * df.petal_length
Analysis: Feature engineering¶
Elementwise operations are very useful in machine learning, especially for feature engineering.
Feature engineering is the process of creating new features (model input variables) from existing ones.
Let's first use our toy_df to illustrate the concept.
In the Iris dataset, we have petal width and length, but what if we wanted to know petal area? Well, we can create a new petal_area feature.
**First, display the two columns of
petal_width and petal_length in toy_df.**
* **Tip:** You can index a DataFrame using a list of column names too, like so:
df[['column_1', 'column_2']]
# Display petal_width and petal_length
toy_df[['petal_width', 'petal_length']]
| petal_width | petal_length | |
|---|---|---|
| 0 | 0.2 | 1.4 |
| 1 | 0.2 | 1.4 |
| 2 | 0.2 | 1.3 |
| 3 | 0.2 | 1.5 |
| 4 | 0.2 | 1.4 |
| 145 | 2.3 | 5.2 |
| 146 | 1.9 | 5.0 |
| 147 | 2.0 | 5.2 |
| 148 | 2.3 | 5.4 |
| 149 | 1.8 | 5.1 |
Next, create a new petal_area feature in toy_df.
We multiply the petal_width column by the petal_length column.
# Create a new petal_area column
toy_df['petal_area'] = toy_df.petal_width * toy_df.petal_length
# Display toy_df
toy_df
| sepal_length | sepal_width | petal_length | petal_width | species | petal_area | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa | 0.28 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa | 0.28 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa | 0.26 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa | 0.30 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa | 0.28 |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | virginica | 11.96 |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | virginica | 9.50 |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | virginica | 10.40 |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica | 12.42 |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | virginica | 9.18 |
By creating a petal_area feature, it's now much easier to see that virginica flowers have significantly larger petals than setosa flowers do!
Often, by creating new features, you can learn more about the data (and improve your machine learning models).
Boolean masks allow filtering¶
Boolean masks¶
Boolean masks are list-like sequences of True/False (boolean) values. These allow you to filter DataFrames in enlightening ways.
"List-like" sequences include:
- lists
- NumPy arrays
- Pandas Series
For example, both of these are valid boolean masks:
list_mask = [True, True, False, True]
series_mask = pd.Series([True, True, False, True])
print( series_mask )
0 True 1 True 2 False 3 True dtype: bool
Conditional boolean masks¶
Often, the more useful way to create a boolean mask is by directly applying a conditional statement to another Pandas Series.
For example, let's say we had the following series:
example_series = pd.Series([10, 5, -3, 2])
print( example_series )
0 10 1 5 2 -3 3 2 dtype: int64
If we wanted to create a boolean mask for all of the positive values in example_series, we can do so like this:
# Create boolean mask from a condition
series_mask = example_series > 0
print( series_mask )
0 True 1 True 2 False 3 True dtype: bool
Filtering¶
Now, we can actually use that boolean mask to filter our Series and keep only the positive observations.
For example:
# Keep only True values from the boolean mask
example_series[series_mask]
0 10 1 5 3 2 dtype: int64
Inverted filtering¶
Or, by using the tilde ~ operator (called the invert operator), we can filter our Series and keep only non-positive observations.
This is equivalent to keeping only the False values from our boolean mask.
For example:
# Keep only False values from the boolean mask
example_series[~series_mask]
2 -3 dtype: int64
Filtering rows¶
# Display [i.e., index] observations where petal_area > 14
df[df.petal_area > 14]
| sepal_length | sepal_width | petal_length | petal_width | species | petal_area | |
|---|---|---|---|---|---|---|
| 100 | 6.3 | 3.3 | 6.0 | 2.5 | virginica | 15.00 |
| 109 | 7.2 | 3.6 | 6.1 | 2.5 | virginica | 15.25 |
| 117 | 7.7 | 3.8 | 6.7 | 2.2 | virginica | 14.74 |
| 118 | 7.7 | 2.6 | 6.9 | 2.3 | virginica | 15.87 |
| 135 | 7.7 | 3.0 | 6.1 | 2.3 | virginica | 14.03 |
| 144 | 6.7 | 3.3 | 5.7 | 2.5 | virginica | 14.25 |
Indicator variables¶
Indicator variables are variables that can take on one of two values:
- 1 if a condition is met.
- 0 if a condition is not met.
In machine learning, we want the values to be 1/0 instead of True/False because our algorithms will require numeric inputs.
Fortunately, you can convert a boolean mask into 1/0's using .astype(int):
# Example boolean Series
example_mask = pd.Series([True, False, False, True, False])
# Convert boolean Series into 1/0
print( example_mask.astype(int) )
0 1 1 0 2 0 3 1 4 0 dtype: int64
Thus, if we want to create an indicator variable for petal_area > 14, we can do so in one line of code:
# Create indicator variable for petal_area > 14
df['giant'] = (df.petal_area > 14).astype(int)
df.head()
| sepal_length | sepal_width | petal_length | petal_width | species | petal_area | giant | |
|---|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa | 0.28 | 0 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa | 0.28 | 0 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa | 0.26 | 0 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa | 0.30 | 0 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa | 0.28 | 0 |
Multiple masks¶
DataFrames can be indexed by any number of masks, separated by an & operator.
Let's say we wanted to see only versicolor and virginica flowers with sepal_length > 3.2.
- We can use the
.isin()function to see if species is either versicolor or virginica. - To keep our code clean, if we have multiple masks, we might set them to separate variables.
- We'll then combine the masks using the & operator.
# Versicolor or virginica
species_mask = df.species.isin(['versicolor', 'virginica'])
# Sepal width > 3.2
sepal_width_mask = df.sepal_width > 3.2
# Index with both masks
df[species_mask & sepal_width_mask]
| sepal_length | sepal_width | petal_length | petal_width | species | petal_area | giant | |
|---|---|---|---|---|---|---|---|
| 56 | 6.3 | 3.3 | 4.7 | 1.6 | versicolor | 7.52 | 0 |
| 85 | 6.0 | 3.4 | 4.5 | 1.6 | versicolor | 7.20 | 0 |
| 100 | 6.3 | 3.3 | 6.0 | 2.5 | virginica | 15.00 | 1 |
| 109 | 7.2 | 3.6 | 6.1 | 2.5 | virginica | 15.25 | 1 |
| 117 | 7.7 | 3.8 | 6.7 | 2.2 | virginica | 14.74 | 1 |
| 124 | 6.7 | 3.3 | 5.7 | 2.1 | virginica | 11.97 | 0 |
| 131 | 7.9 | 3.8 | 6.4 | 2.0 | virginica | 12.80 | 0 |
| 136 | 6.3 | 3.4 | 5.6 | 2.4 | virginica | 13.44 | 0 |
| 144 | 6.7 | 3.3 | 5.7 | 2.5 | virginica | 14.25 | 1 |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica | 12.42 | 0 |
Analaysis: Advanced filtering with boolean masks¶
Again, we'll use the toy_df to really drive home these concepts.
Let's say we wanted to display observations where petal_area > 10 and sepal_width > 3.
**First, display
toy_df again.**
# Display toy_df
toy_df
| sepal_length | sepal_width | petal_length | petal_width | species | petal_area | |
|---|---|---|---|---|---|---|
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa | 0.28 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa | 0.28 |
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa | 0.26 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa | 0.30 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa | 0.28 |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | virginica | 11.96 |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | virginica | 9.50 |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | virginica | 10.40 |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica | 12.42 |
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | virginica | 9.18 |
Create a boolean mask for petal_area > 10.
# Mask for petal_area > 10
petal_area_mask = toy_df.petal_area > 10
# Display petal_area_mask
petal_area_mask
0 False 1 False 2 False 3 False 4 False 145 True 146 False 147 True 148 True 149 False Name: petal_area, dtype: bool
Next, create a boolean mask for sepal_width > 3.
# Mask for sepal_width > 3
sepal_width_mask = toy_df.sepal_width > 3
# Display sepal_width_mask
sepal_width_mask
0 True 1 False 2 True 3 True 4 True 145 False 146 False 147 False 148 True 149 False Name: sepal_width, dtype: bool
Next, display the two masks combined using the & operator.
# Display both masks, combined
petal_area_mask & sepal_width_mask
0 False 1 False 2 False 3 False 4 False 145 False 146 False 147 False 148 True 149 False dtype: bool
Finally, select the observations from toy_df where both conditions are met.
# Index with both masks
toy_df[petal_area_mask & sepal_width_mask]
| sepal_length | sepal_width | petal_length | petal_width | species | petal_area | |
|---|---|---|---|---|---|---|
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica | 12.42 |
Groupby allow segmentation¶
Groupby¶
The .groupby() function allows you to segment and summarize data across different classes.
For example, let's say we wanted to find the average measurements for each of the 3 species of Iris flowers.
- First, take our DataFrame and group by species:
df.groupby('species') - Then, simply take the mean by column:
df.groupby('species').mean()
Here's how the output looks:
# Display average measurements for each species
df.groupby('species').mean()
| sepal_length | sepal_width | petal_length | petal_width | petal_area | giant | |
|---|---|---|---|---|---|---|
| species | ||||||
| setosa | 5.006 | 3.428 | 1.462 | 0.246 | 0.3656 | 0.00 |
| versicolor | 5.936 | 2.770 | 4.260 | 1.326 | 5.7204 | 0.00 |
| virginica | 6.588 | 2.974 | 5.552 | 2.026 | 11.2962 | 0.12 |
Agg¶
Finally, what if we wanted to display multiple pieces of information (or aggregations)?
Well, we can use the .agg() function and pass in a list of aggregations, like so:
# Display min, median, max measurements for each species
df.groupby('species').agg(['min', 'median', 'max'])
| sepal_length | sepal_width | petal_length | petal_width | petal_area | giant | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| min | median | max | min | median | max | min | median | max | min | median | max | min | median | max | min | median | max | |
| species | ||||||||||||||||||
| setosa | 4.3 | 5.0 | 5.8 | 2.3 | 3.4 | 4.4 | 1.0 | 1.50 | 1.9 | 0.1 | 0.2 | 0.6 | 0.11 | 0.300 | 0.96 | 0 | 0 | 0 |
| versicolor | 4.9 | 5.9 | 7.0 | 2.0 | 2.8 | 3.4 | 3.0 | 4.35 | 5.1 | 1.0 | 1.3 | 1.8 | 3.30 | 5.615 | 8.64 | 0 | 0 | 0 |
| virginica | 4.9 | 6.5 | 7.9 | 2.2 | 3.0 | 3.8 | 4.5 | 5.55 | 6.9 | 1.4 | 2.0 | 2.5 | 7.50 | 11.445 | 15.87 | 0 | 0 | 1 |
Analysis: aggregating using groupby¶
Now, armed with the power of groupby, we're almost ready to return to Casey and share what we've learned about Iris flowers! But before we do, let's just bring back our toy_df for one last hoorah, just to make sure we know what's going on under the hood.
Let's calculate the median
petal_area for each species.
* Since toy_df is small, we can do this manually as well and check to make sure the values are correct.
**First, let's manually calculate the median
petal_area for the virginica flowers in our toy_df.**
# Display all 'virginica' species; values sorted by petal_area (both work)
# Way 1
toy_df[toy_df.species.isin(['virginica'])].sort_values('petal_area')
# Way 2
toy_df[toy_df.species == 'virginica'].sort_values(by='petal_area')
| sepal_length | sepal_width | petal_length | petal_width | species | petal_area | |
|---|---|---|---|---|---|---|
| 149 | 5.9 | 3.0 | 5.1 | 1.8 | virginica | 9.18 |
| 146 | 6.3 | 2.5 | 5.0 | 1.9 | virginica | 9.50 |
| 147 | 6.5 | 3.0 | 5.2 | 2.0 | virginica | 10.40 |
| 145 | 6.7 | 3.0 | 5.2 | 2.3 | virginica | 11.96 |
| 148 | 6.2 | 3.4 | 5.4 | 2.3 | virginica | 12.42 |
Based on the output above, what's median petal_area for the virginica species?
**Next, let's manually calculate the median
petal_area for the setosa flowers in our toy_df.**
# Display all 'setosa' species (both ways work the same)
# Way 1
toy_df[toy_df.species.isin(['setosa'])].sort_values('petal_area')
# Way 2
toy_df[toy_df.species == 'setosa'].sort_values(by='petal_area')
| sepal_length | sepal_width | petal_length | petal_width | species | petal_area | |
|---|---|---|---|---|---|---|
| 2 | 4.7 | 3.2 | 1.3 | 0.2 | setosa | 0.26 |
| 0 | 5.1 | 3.5 | 1.4 | 0.2 | setosa | 0.28 |
| 1 | 4.9 | 3.0 | 1.4 | 0.2 | setosa | 0.28 |
| 4 | 5.0 | 3.6 | 1.4 | 0.2 | setosa | 0.28 |
| 3 | 4.6 | 3.1 | 1.5 | 0.2 | setosa | 0.30 |
Based on the output above, what's median petal_area for the setosa species?
**Finally let's calculate the median values using a
.groupby().**
* Should get the same result!
# Median petal_area in toy_df of all features
toy_df.groupby('species').median()
| sepal_length | sepal_width | petal_length | petal_width | petal_area | |
|---|---|---|---|---|---|
| species | |||||
| setosa | 4.9 | 3.2 | 1.4 | 0.2 | 0.28 |
| virginica | 6.3 | 3.0 | 5.2 | 2.0 | 10.40 |