Classification of Italian Wines¶

In this notebook we will be using supervised learning to classify Italian wines. The question is: Can we teach a machine to figure out which type of wine an obseration belongs to?
We will work with a famous but small dataset that can be found here (along more informaion). The data is clean, contains only numerical and no missing values. We will not do any EDA but only focus on prediction. The only preprocessing step will be standardization of the physiochemical variables.
We will be using Pandas and Scikit-Learn which are both parts of the Anaconda distribution.
# Download the dateset using WGET.
# If this is not possible, then just paste the URL in your browser and download
# the file, or if you use GithubDesktop then it should be in the folder
# after a pull.
!wget https://cdn.rawgit.com/SDS-AAU/M1-2018/182abaa2/data/wine.csv
Redirecting output to ‘wget-log.1’.
# Importing the libraries
import numpy as np # for working with arrays
np.set_printoptions(suppress=True) # not a must but nice to avoid scientific notation
import pandas as pd # as usual for handling dataframes
pd.options.display.float_format = '{:.4f}'.format #same for pandas to turn off scientific notation
# Importing the dataset
dataset = pd.read_csv('wine.csv')
# Quick check of the dataframe proportions
dataset.shape
(178, 15)
# Checking the first 5 rows to get familiar with the data
dataset.head()
| class_label | class_name | alcohol | malic_acid | ash | alcalinity_of_ash | magnesium | total_phenols | flavanoids | nonflavanoid_phenols | proanthocyanins | color_intensity | hue | od280 | proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | Barolo | 14.2300 | 1.7100 | 2.4300 | 15.6000 | 127 | 2.8000 | 3.0600 | 0.2800 | 2.2900 | 5.6400 | 1.0400 | 3.9200 | 1065 |
| 1 | 1 | Barolo | 13.2000 | 1.7800 | 2.1400 | 11.2000 | 100 | 2.6500 | 2.7600 | 0.2600 | 1.2800 | 4.3800 | 1.0500 | 3.4000 | 1050 |
| 2 | 1 | Barolo | 13.1600 | 2.3600 | 2.6700 | 18.6000 | 101 | 2.8000 | 3.2400 | 0.3000 | 2.8100 | 5.6800 | 1.0300 | 3.1700 | 1185 |
| 3 | 1 | Barolo | 14.3700 | 1.9500 | 2.5000 | 16.8000 | 113 | 3.8500 | 3.4900 | 0.2400 | 2.1800 | 7.8000 | 0.8600 | 3.4500 | 1480 |
| 4 | 1 | Barolo | 13.2400 | 2.5900 | 2.8700 | 21.0000 | 118 | 2.8000 | 2.6900 | 0.3900 | 1.8200 | 4.3200 | 1.0400 | 2.9300 | 735 |
# Getting basic descriptives for all nummerical variables
dataset.describe()
| class_label | alcohol | malic_acid | ash | alcalinity_of_ash | magnesium | total_phenols | flavanoids | nonflavanoid_phenols | proanthocyanins | color_intensity | hue | od280 | proline | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 |
| mean | 1.9382 | 13.0006 | 2.3363 | 2.3665 | 19.4949 | 99.7416 | 2.2951 | 2.0293 | 0.3619 | 1.5909 | 5.0581 | 0.9574 | 2.6117 | 746.8933 |
| std | 0.7750 | 0.8118 | 1.1171 | 0.2743 | 3.3396 | 14.2825 | 0.6259 | 0.9989 | 0.1245 | 0.5724 | 2.3183 | 0.2286 | 0.7100 | 314.9075 |
| min | 1.0000 | 11.0300 | 0.7400 | 1.3600 | 10.6000 | 70.0000 | 0.9800 | 0.3400 | 0.1300 | 0.4100 | 1.2800 | 0.4800 | 1.2700 | 278.0000 |
| 25% | 1.0000 | 12.3625 | 1.6025 | 2.2100 | 17.2000 | 88.0000 | 1.7425 | 1.2050 | 0.2700 | 1.2500 | 3.2200 | 0.7825 | 1.9375 | 500.5000 |
| 50% | 2.0000 | 13.0500 | 1.8650 | 2.3600 | 19.5000 | 98.0000 | 2.3550 | 2.1350 | 0.3400 | 1.5550 | 4.6900 | 0.9650 | 2.7800 | 673.5000 |
| 75% | 3.0000 | 13.6775 | 3.0825 | 2.5575 | 21.5000 | 107.0000 | 2.8000 | 2.8750 | 0.4375 | 1.9500 | 6.2000 | 1.1200 | 3.1700 | 985.0000 |
| max | 3.0000 | 14.8300 | 5.8000 | 3.2300 | 30.0000 | 162.0000 | 3.8800 | 5.0800 | 0.6600 | 3.5800 | 13.0000 | 1.7100 | 4.0000 | 1680.0000 |
We can see here that means and spread (standard deviation) of the features is very different and thus we will need to standardize the dataset.
"As a rule of thumb I’d say: When in doubt, just standardize the data, it shouldn’t hurt."" Sebastian Raschka
# Selecting the relevant data
# using the iloc selector allows to grab a range 2-15 of columns
# withouth having to call their names. That's practical
# Also, we ask for values only, as we are going to pass the data into
# the ML algorithms in the form of arrays rather than pandas DFs
X = dataset.iloc[:, 2:15].values
y = dataset.iloc[:, 1].values
Yes, there is a class_lable in the dataset but for the sake of learning and because it is very simple, we are going to construct our class_lables on our own. For this we will use the LabelEncoder from Scikit-Learn. Note that in contrast to Pandas, the Scikit-Learn is more of a (HUGE!!!) Library where you have to import different functionalities separately. You can find an index of all classes here.
# Encoding categorical data
from sklearn.preprocessing import LabelEncoder
Classes such as the LabelEncoder or any modely type that you import have several parameters that can (but don't have to be) specified. Also, you are usually fitting them to some data first before performind transformations. Thus, they are cutom-made for each use case and therefore you will need to define an encoder object from the imported class. This is a general philosophy behind all Scikit-Learn classes. The good news: The syntax is the same across all classes.
Below we first define a labelencoder_y and then use the fit_transform method (we could also first use fit and then transform) to turn our wine-type names into numbers.
# From labels to numbers
labelencoder_y = LabelEncoder()
y = labelencoder_y.fit_transform(y)
As you have seen from the descriptives above our variables lie on very different scales. Therefore, we will standardize them before going further. The procedure using the StandardScaleris exactly the same as before with the label encoder.
This scaling will for each value substract the mean (of the column) and devide it by the standard deviation, thus bringing them all on the same scale with a mean of 0 and a standard deviation of 1.
# Feature scaling
from sklearn.preprocessing import StandardScaler
scaler = StandardScaler()
X = scaler.fit_transform(X)
# We can check our transform data using pandas describe
pd.DataFrame(X).describe()
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 | 178.0000 |
| mean | -0.0000 | -0.0000 | -0.0000 | -0.0000 | -0.0000 | 0.0000 | -0.0000 | 0.0000 | -0.0000 | 0.0000 | 0.0000 | 0.0000 | -0.0000 |
| std | 1.0028 | 1.0028 | 1.0028 | 1.0028 | 1.0028 | 1.0028 | 1.0028 | 1.0028 | 1.0028 | 1.0028 | 1.0028 | 1.0028 | 1.0028 |
| min | -2.4342 | -1.4330 | -3.6792 | -2.6710 | -2.0883 | -2.1072 | -1.6960 | -1.8682 | -2.0690 | -1.6343 | -2.0947 | -1.8951 | -1.4932 |
| 25% | -0.7882 | -0.6587 | -0.5721 | -0.6891 | -0.8244 | -0.8855 | -0.8275 | -0.7401 | -0.5973 | -0.7951 | -0.7676 | -0.9522 | -0.7846 |
| 50% | 0.0610 | -0.4231 | -0.0238 | 0.0015 | -0.1223 | 0.0960 | 0.1061 | -0.1761 | -0.0629 | -0.1592 | 0.0331 | 0.2377 | -0.2337 |
| 75% | 0.8361 | 0.6698 | 0.6981 | 0.6021 | 0.5096 | 0.8090 | 0.8491 | 0.6095 | 0.6292 | 0.4940 | 0.7132 | 0.7886 | 0.7582 |
| max | 2.2598 | 3.1092 | 3.1563 | 3.1545 | 4.3714 | 2.5395 | 3.0628 | 2.4024 | 3.4851 | 3.4354 | 3.3017 | 1.9609 | 2.9715 |
In the next step we split the data into a training and a test-set. Very often you will see a split of 80/20 %
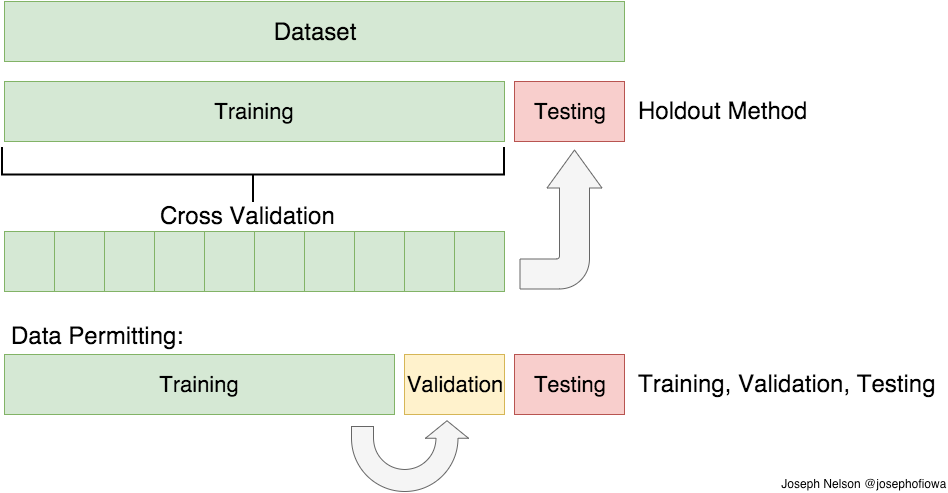
80% of the data will be used to fit a model, while we will keep 20% of the data for testing the models performance.
The train_test_split class takes 4 parameters: (X, y, test_size = 0.2, random_state = 21)
- Input matrix: X
- Output matrix: y
- The test size: We take 20%
- A random state (optional): Some number for the random generator that will shuffle the values*
*The whole random state thing is mostly for easier reproducibility and can also be let our.
# Splitting the dataset into the Training set and Test set
from sklearn.model_selection import train_test_split
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size = 0.2, random_state = 21)

Now it's time for the model to meet the wine data.
We will be using 3 different models. The reason why we use 3 models is because, it is nice to see how easy it is to switch them aroun to experiment what works best. Since we can calculate an (kind of) objective quality measure, it is easy to compare and evaluate them agains each other.
- Logistic Regression
- Suport Vector Classifier
- Random Forest Classifier
Remember that this is a classification problem rather than a regression. The models will be estimating probabilities for some class vs. other classes.
# We first import and train a Logistic Regression
from sklearn.linear_model import LogisticRegression
classifier = LogisticRegression(random_state = 22)
classifier.fit(X_train, y_train)
# After training the model we should jump further down (over the next 2 models)
# To evaluate the results
LogisticRegression(C=1.0, class_weight=None, dual=False, fit_intercept=True,
intercept_scaling=1, max_iter=100, multi_class='ovr', n_jobs=1,
penalty='l2', random_state=22, solver='liblinear', tol=0.0001,
verbose=0, warm_start=False)
# Fitting Random Forest Classification to the Training set
from sklearn.ensemble import RandomForestClassifier
classifier = RandomForestClassifier(n_estimators = 50, criterion = 'entropy', random_state = 22)
classifier.fit(X_train, y_train)
RandomForestClassifier(bootstrap=True, class_weight=None, criterion='entropy',
max_depth=None, max_features='auto', max_leaf_nodes=None,
min_impurity_decrease=0.0, min_impurity_split=None,
min_samples_leaf=1, min_samples_split=2,
min_weight_fraction_leaf=0.0, n_estimators=50, n_jobs=1,
oob_score=False, random_state=22, verbose=0, warm_start=False)
# Finally we train a Support Vector Classifier
from sklearn.svm import SVC
classifier = SVC(kernel = 'linear', random_state = 21)
classifier.fit(X_train, y_train)
SVC(C=1.0, cache_size=200, class_weight=None, coef0=0.0, decision_function_shape='ovr', degree=3, gamma='auto', kernel='linear', max_iter=-1, probability=False, random_state=21, shrinking=True, tol=0.001, verbose=False)
Perhaps this time the algorithm was just lucky because of a random allocation of the data in the train-test split. To make sure which model is the most accurate, we can run a k-Fold Cross Validation deviding x_train into (here) 10 parts, training on 9 and testing on 1. This will be done 10 times, every time measuring the accuracy and finally returning the average accuracy.

# Applying k-Fold Cross Validation
from sklearn.model_selection import cross_val_score
accuracies = cross_val_score(estimator = classifier, X = X_train, y = y_train, cv = 5)
print(accuracies.mean())
print(accuracies.std())
0.9856960408684546 0.017537311768860152
Now that we fitted or trained a model we need to figure out how well it performes. This approach to evaluation is very different from what many of you are used to from econometrics.
Here we are not interested in a model summary table, rather we will be exploring predictive performance. In the next cell we ask the classifier object (our trained model) to gives us predictions for data it never has seen before.
Then we will compare the predictions made against the real-world values that we actually know.
# Predicting the Test set results
y_pred = classifier.predict(X_test)
# Making a classification report
from sklearn.metrics import classification_report
cm = classification_report(y_test, y_pred)
print(cm)
precision recall f1-score support
0 0.92 1.00 0.96 11
1 1.00 1.00 1.00 15
2 1.00 0.90 0.95 10
avg / total 0.97 0.97 0.97 36
There is also a slightly more intuitive way to evaluate our predictions in the case of a multiclass-classification where we cannot just create a confusion-matrix. What we can do is using pandas to crosstabulate our real against our predicted wines.
To get the wine names, we will use the inverse_transform function of our labelencoder
# Transforming nummerical labels to wine types
true_wines = labelencoder_y.inverse_transform(y_test)
predicted_wines = labelencoder_y.inverse_transform(y_pred)
/usr/local/lib/python3.6/dist-packages/sklearn/preprocessing/label.py:151: DeprecationWarning: The truth value of an empty array is ambiguous. Returning False, but in future this will result in an error. Use `array.size > 0` to check that an array is not empty. if diff: /usr/local/lib/python3.6/dist-packages/sklearn/preprocessing/label.py:151: DeprecationWarning: The truth value of an empty array is ambiguous. Returning False, but in future this will result in an error. Use `array.size > 0` to check that an array is not empty. if diff:
# Creating a pandas DataFrame and cross-tabulation
df = pd.DataFrame({'true_wines': true_wines, 'predicted_wines': predicted_wines})
pd.crosstab(df.true_wines, df.predicted_wines)
| predicted_wines | Barbera | Barolo | Grignolino |
|---|---|---|---|
| true_wines | |||
| Barbera | 11 | 0 | 0 |
| Barolo | 0 | 15 | 0 |
| Grignolino | 1 | 0 | 9 |
But is that not the same as PCA or soe other kind of clustering?
Well, let's try to use unsupervised learning on the same data-set. We will be using KMeans (because it is simple and nice for illustration)
Just as before, we import a model class, define a model object and fit it. Same 3 steps as before.
# We import KMeans and creade a model object (we know that there are 3 wines...kind of cheating)
from sklearn.cluster import KMeans
model = KMeans(n_clusters = 3)
# Fitting the model is super easy, jsut one line
model.fit(X_train)
KMeans(algorithm='auto', copy_x=True, init='k-means++', max_iter=300,
n_clusters=3, n_init=10, n_jobs=1, precompute_distances='auto',
random_state=None, tol=0.0001, verbose=0)
# Prediction is easy, too
predicted_wine_clusters = model.predict(X_train)
predicted_new_wine_clusters = model.predict(X_test)
Note that the clustering model never met any y-values - only X values
# Quick print out of the labels
predicted_wine_clusters
array([0, 2, 0, 1, 2, 2, 1, 2, 1, 1, 0, 0, 2, 2, 1, 0, 0, 0, 2, 2, 2, 2,
1, 0, 0, 1, 0, 2, 2, 0, 1, 0, 0, 0, 0, 1, 2, 2, 1, 1, 1, 1, 1, 1,
1, 2, 0, 1, 2, 1, 0, 2, 2, 0, 2, 2, 2, 2, 2, 0, 1, 1, 2, 1, 2, 1,
1, 2, 1, 1, 0, 2, 1, 0, 2, 2, 0, 1, 0, 1, 1, 1, 2, 0, 2, 2, 1, 1,
2, 2, 0, 1, 1, 1, 2, 2, 2, 2, 0, 0, 0, 1, 2, 0, 0, 1, 0, 2, 2, 0,
1, 2, 0, 1, 2, 1, 0, 1, 2, 1, 1, 0, 1, 1, 0, 2, 0, 2, 2, 2, 0, 2,
0, 2, 2, 2, 0, 2, 2, 1, 1, 1], dtype=int32)
# Transforming nummerical labels to wine types
true_wines = labelencoder_y.inverse_transform(y_train)
df = pd.DataFrame({'true_wines': true_wines, 'predicted_wines': predicted_wine_clusters})
pd.crosstab(df.true_wines, df.predicted_wines)
/usr/local/lib/python3.6/dist-packages/sklearn/preprocessing/label.py:151: DeprecationWarning: The truth value of an empty array is ambiguous. Returning False, but in future this will result in an error. Use `array.size > 0` to check that an array is not empty. if diff:
| predicted_wines | 0 | 1 | 2 |
|---|---|---|---|
| true_wines | |||
| Barbera | 37 | 0 | 0 |
| Barolo | 0 | 44 | 0 |
| Grignolino | 3 | 4 | 54 |