Literature / Useful References¶
Books¶
- Jean Claude, Morphometry with R
- Online through ETHZ
- Buy it
- John C. Russ, “The Image Processing Handbook”,(Boca Raton, CRC Press)
- Available online within domain ethz.ch (or proxy.ethz.ch / public VPN)
Papers / Sites¶
- Thickness
- [1] Hildebrand, T., & Ruegsegger, P. (1997). A new method for the model-independent assessment of thickness in three-dimensional images. Journal of Microscopy, 185(1), 67–75. doi:10.1046/j.1365-2818.1997.1340694.x
- Curvature
- http://mathworld.wolfram.com/MeanCurvature.html
- [2] "Computation of Surface Curvature from Range Images Using Geometrically Intrinsic Weights"*, T. Kurita and P. Boulanger, 1992.
- http://radiomics.io
Outline¶
- Motivation (Why and How?)
- What are Distance Maps?
- What are thickness maps?
- Characteristic Shapes
- Texture Analysis
Learning Objectives¶
Motivation (Why and How?)¶
How can we measure sizes in complicated objects?
How do we measure sizes relavant for diffusion or other local processes?
How do we investigate surfaces in more detail and their shape?
How can we compare shape of complex objects when they grow?
Are there characteristic shape metrics?
- How to we quantify patterns inside images?
- How can compare between different patterns?
- How can we quantify as radiologists say looking evil?
Distance Maps: What are they¶
%matplotlib inline
import numpy as np
import skimage.transform
from scipy import ndimage
import matplotlib.pyplot as plt
import seaborn as sns
import os
def generate_dot_image(size=100, rad_a=1, rad_b=None):
xx, yy = np.meshgrid(np.linspace(-1, 1, size), np.linspace(-1, 1, size))
if rad_b is None:
rad_b = rad_a
return np.sqrt(np.square(xx/rad_a)+np.square(yy/rad_b)) <= 1.0
img_bw = generate_dot_image(10, 0.8)
sns.heatmap(img_bw, annot=True,
fmt="d", cmap='gray')
<matplotlib.axes._subplots.AxesSubplot at 0x221cd8c2d30>
A map (or image) of distances. Each point in the map is the distance that point is from a given feature of interest (surface of an object, ROI, center of object, etc)
help(ndimage.distance_transform_edt)
Help on function distance_transform_edt in module scipy.ndimage.morphology:
distance_transform_edt(input, sampling=None, return_distances=True, return_indices=False, distances=None, indices=None)
Exact euclidean distance transform.
In addition to the distance transform, the feature transform can
be calculated. In this case the index of the closest background
element is returned along the first axis of the result.
Parameters
----------
input : array_like
Input data to transform. Can be any type but will be converted
into binary: 1 wherever input equates to True, 0 elsewhere.
sampling : float or int, or sequence of same, optional
Spacing of elements along each dimension. If a sequence, must be of
length equal to the input rank; if a single number, this is used for
all axes. If not specified, a grid spacing of unity is implied.
return_distances : bool, optional
Whether to return distance matrix. At least one of
return_distances/return_indices must be True. Default is True.
return_indices : bool, optional
Whether to return indices matrix. Default is False.
distances : ndarray, optional
Used for output of distance array, must be of type float64.
indices : ndarray, optional
Used for output of indices, must be of type int32.
Returns
-------
distance_transform_edt : ndarray or list of ndarrays
Either distance matrix, index matrix, or a list of the two,
depending on `return_x` flags and `distance` and `indices`
input parameters.
Notes
-----
The euclidean distance transform gives values of the euclidean
distance::
n
y_i = sqrt(sum (x[i]-b[i])**2)
i
where b[i] is the background point (value 0) with the smallest
Euclidean distance to input points x[i], and n is the
number of dimensions.
Examples
--------
>>> from scipy import ndimage
>>> a = np.array(([0,1,1,1,1],
... [0,0,1,1,1],
... [0,1,1,1,1],
... [0,1,1,1,0],
... [0,1,1,0,0]))
>>> ndimage.distance_transform_edt(a)
array([[ 0. , 1. , 1.4142, 2.2361, 3. ],
[ 0. , 0. , 1. , 2. , 2. ],
[ 0. , 1. , 1.4142, 1.4142, 1. ],
[ 0. , 1. , 1.4142, 1. , 0. ],
[ 0. , 1. , 1. , 0. , 0. ]])
With a sampling of 2 units along x, 1 along y:
>>> ndimage.distance_transform_edt(a, sampling=[2,1])
array([[ 0. , 1. , 2. , 2.8284, 3.6056],
[ 0. , 0. , 1. , 2. , 3. ],
[ 0. , 1. , 2. , 2.2361, 2. ],
[ 0. , 1. , 2. , 1. , 0. ],
[ 0. , 1. , 1. , 0. , 0. ]])
Asking for indices as well:
>>> edt, inds = ndimage.distance_transform_edt(a, return_indices=True)
>>> inds
array([[[0, 0, 1, 1, 3],
[1, 1, 1, 1, 3],
[2, 2, 1, 3, 3],
[3, 3, 4, 4, 3],
[4, 4, 4, 4, 4]],
[[0, 0, 1, 1, 4],
[0, 1, 1, 1, 4],
[0, 0, 1, 4, 4],
[0, 0, 3, 3, 4],
[0, 0, 3, 3, 4]]])
With arrays provided for inplace outputs:
>>> indices = np.zeros(((np.ndim(a),) + a.shape), dtype=np.int32)
>>> ndimage.distance_transform_edt(a, return_indices=True, indices=indices)
array([[ 0. , 1. , 1.4142, 2.2361, 3. ],
[ 0. , 0. , 1. , 2. , 2. ],
[ 0. , 1. , 1.4142, 1.4142, 1. ],
[ 0. , 1. , 1.4142, 1. , 0. ],
[ 0. , 1. , 1. , 0. , 0. ]])
>>> indices
array([[[0, 0, 1, 1, 3],
[1, 1, 1, 1, 3],
[2, 2, 1, 3, 3],
[3, 3, 4, 4, 3],
[4, 4, 4, 4, 4]],
[[0, 0, 1, 1, 4],
[0, 1, 1, 1, 4],
[0, 0, 1, 4, 4],
[0, 0, 3, 3, 4],
[0, 0, 3, 3, 4]]])
dmap = ndimage.distance_transform_edt(img_bw)
sns.heatmap(dmap, annot=True,
fmt="2.1f", cmap='nipy_spectral')
<matplotlib.axes._subplots.AxesSubplot at 0x221d0a4ed68>
from timeit import timeit
dmap_list = [ndimage.distance_transform_edt,
ndimage.distance_transform_bf, ndimage.distance_transform_cdt]
fig, m_axs = plt.subplots(1, len(dmap_list), figsize=(20, 6))
for dmap_func, c_ax in zip(dmap_list, m_axs):
ms_time = timeit(lambda: dmap_func(img_bw), number=10000)/10000*1e6
dmap = dmap_func(img_bw)
sns.heatmap(dmap, annot=True,
fmt="2.1f", cmap='nipy_spectral', ax=c_ax, cbar=False)
c_ax.set_title('{}\n{} - {} us'.format(dmap_func.__doc__.split('\n')[1].strip(),
dmap_func.__name__,
'%2.2f' % ms_time))
Why does speed matter?¶
As for the question why speed matters, for small images clearly the more efficient approaches don't make much of a difference, but when we start to look at data from a Synchrotron measurement 2160x2560x2560 (SLS) this becomes much more important
img_bw = generate_dot_image(512, 0.75)
sls_fact = 2160*2560*2560/np.prod(img_bw.shape)
dmap_list = [ndimage.distance_transform_edt,
ndimage.distance_transform_bf, ndimage.distance_transform_cdt]
fig, m_axs = plt.subplots(1, len(dmap_list), figsize=(20, 6))
for dmap_func, c_ax in zip(dmap_list, m_axs):
ms_time = timeit(lambda: dmap_func(img_bw), number=1)*1e3
dmap = dmap_func(img_bw)
c_ax.matshow(dmap, cmap='nipy_spectral')
c_ax.set_title('{}\n{} : {} ms\n For SLS scan {} h'.format(dmap_func.__doc__.split('\n')[1].strip(),
dmap_func.__name__,
'%2.2f' % ms_time,
'%2.2f' % (ms_time*sls_fact/3600/1e3)))
Definition¶
If we start with an image as a collection of points divided into two categories
- Im(x,y)= {Foreground, Background}
- We can define a distance map operator (dist) that transforms the image into a distance map
We will use Euclidean distance ||→x−→y|| for this class but there are other metrics which make sense when dealing with other types of data like Manhattan/City-block or weighted metrics.
def simple_distance_iteration(last_img):
cur_img = last_img.copy()
for x in range(last_img.shape[0]):
for y in range(last_img.shape[1]):
if last_img[x, y] >= 0:
i_xy = last_img[x, y]
for xp in [-1, 0, 1]:
if (x+xp < last_img.shape[0]) and (x+xp >= 0):
for yp in [-1, 0, 1]:
if (y+yp < last_img.shape[1]) and (y+yp >= 0):
p_dist = np.sqrt(np.square(xp)+np.square(yp))
if cur_img[x+xp, y+yp] > (last_img[x, y]+p_dist):
cur_img[x+xp, y+yp] = last_img[x, y]+p_dist
return cur_img
img_bw = generate_dot_image(10, 1.0)
sns.heatmap(img_bw, annot=True,
fmt="d", cmap='gray')
<matplotlib.axes._subplots.AxesSubplot at 0x221d0e244e0>
from matplotlib.animation import FuncAnimation
from IPython.display import HTML
fig, c_ax = plt.subplots(1, 1, figsize=(5, 5), dpi=150)
img_base = (img_bw*255).astype(np.float32)
img_list = [img_base]
for i in range(5):
img_base = simple_distance_iteration(img_base)
img_list += [img_base]
def update_frame(i):
plt.cla()
sns.heatmap(img_list[i],
annot=True,
fmt="2.1f",
cmap='nipy_spectral',
ax=c_ax,
cbar=False,
vmin=0,
vmax=5)
c_ax.set_title('Iteration #{}'.format(i+1))
# write animation frames
anim_code = FuncAnimation(fig,
update_frame,
frames=5,
interval=1000,
repeat_delay=2000).to_html5_video()
plt.close('all')
HTML(anim_code)
Distance Maps: Types¶
Using this rule a distance map can be made for the euclidean metric
Similarly the Manhattan or city block distance metric can be used where the distance is defined as ∑i|→x−→y|i
def simple_distance_iteration(last_img):
cur_img = last_img.copy()
for x in range(last_img.shape[0]):
for y in range(last_img.shape[1]):
if last_img[x, y] >= 0:
i_xy = last_img[x, y]
for xp in [-1, 0, 1]:
if (x+xp < last_img.shape[0]) and (x+xp >= 0):
for yp in [-1, 0, 1]:
if (y+yp < last_img.shape[1]) and (y+yp >= 0):
p_dist = np.abs(xp)+np.abs(yp)
if cur_img[x+xp, y+yp] > (last_img[x, y]+p_dist):
cur_img[x+xp, y+yp] = last_img[x, y]+p_dist
return cur_img
from matplotlib.animation import FuncAnimation
from IPython.display import HTML
fig, c_ax = plt.subplots(1, 1, figsize=(5, 5), dpi=150)
img_base = (img_bw*255).astype(np.float32)
img_list = [img_base]
for i in range(5):
img_base = simple_distance_iteration(img_base)
img_list += [img_base]
def update_frame(i):
plt.cla()
sns.heatmap(img_list[i],
annot=True,
fmt="2.0f",
cmap='nipy_spectral',
ax=c_ax,
cbar=False,
vmin=0,
vmax=5)
c_ax.set_title('Iteration #{}'.format(i+1))
c_ax.set_aspect(1)
# write animation frames
anim_code = FuncAnimation(fig,
update_frame,
frames=5,
interval=1000,
repeat_delay=2000).to_html5_video()
plt.close('all')
HTML(anim_code)
Distance Maps: Precaution¶
The distance map is one of the crictical points where the resolution of the imaging system is important.
- We measure distances computationally in pixels or voxels
- but for them to have a meaning physically they must be converted
- Isotropic imaging (1 μ m x 1μ m x 1 μ m) is fine
Anisotropic¶
Ideally
- as part of filtering, resample and convert to an isotropic scale.
If that is not possible
- custom distance map algorithms which use the side-lengths of the voxels to calculate distance rather than assuming 1x1x1
Distance Maps¶
We can create 2 distance maps
- Foreground → Background
- Information about the objects size and interior
- Background → Foreground
- Information about the distance / space between objects
%matplotlib inline
import numpy as np
import skimage.transform
from scipy import ndimage
import matplotlib.pyplot as plt
import seaborn as sns
import os
def generate_dot_image(size=100, rad_a=1, rad_b=None):
xx, yy = np.meshgrid(np.linspace(-1, 1, size), np.linspace(-1, 1, size))
if rad_b is None:
rad_b = rad_a
return np.sqrt(np.square(xx/rad_a)+np.square(yy/rad_b)) <= 1.0
img_bw = generate_dot_image(10, 0.5, 1.0)
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(20, 6))
sns.heatmap(img_bw, annot=True,
fmt="d", cmap='gray', ax=ax1)
sns.heatmap(ndimage.distance_transform_edt(img_bw),
annot=True,
fmt="2.1f", cmap='nipy_spectral', ax=ax2)
sns.heatmap(ndimage.distance_transform_edt(1-img_bw),
annot=True,
fmt="2.1f", cmap='nipy_spectral', ax=ax3)
<matplotlib.axes._subplots.AxesSubplot at 0x221d1123208>
Distance Map¶
One of the most useful components of the distance map is that it is relatively insensitive to small changes in connectivity.
- Component Labeling would find radically different results for these two images
- One has 4 small circles
- One has 1 big blob
from matplotlib.animation import FuncAnimation
from IPython.display import HTML
fig, m_axs = plt.subplots(2, 3, figsize=(10, 12), dpi=100)
img_list = []
for i in np.linspace(0.6, 1.3, 7):
img_bw = np.concatenate([generate_dot_image(12, 0.7, i),
generate_dot_image(12, 0.6, i)], 0)
img_base = ndimage.distance_transform_edt(img_bw)
img_list += [img_base]
def update_frame(i):
[tax.cla() for tax in m_axs.flatten()]
((c_ax, c_hist, c_dmean), (c_lab, c_labhist, c_lmean)) = m_axs
sns.heatmap(img_list[i],
annot=True,
fmt="2.0f",
cmap='nipy_spectral',
ax=c_ax,
cbar=False,
vmin=0,
vmax=5)
c_ax.set_title('DMap #{}'.format(i+1))
c_hist.hist(img_list[i].ravel(),
np.linspace(0, 3, 6))
c_hist.set_title('Mean: %2.2f\nStd: %2.2f' % (np.mean(img_list[i][img_list[i] > 0]),
np.std(img_list[i][img_list[i] > 0])))
c_dmean.plot(range(i+1), [np.mean(img_list[k][img_list[k] > 0])
for k in range(i+1)], 'r+-')
c_dmean.set_ylim(0, 2)
c_dmean.set_title('Avergage Distance')
lab_img = ndimage.label(img_list[i] > 0)[0]
sns.heatmap(lab_img,
annot=True,
fmt="d",
cmap='nipy_spectral',
ax=c_lab,
cbar=False,
vmin=0,
vmax=2)
c_lab.set_title('Component Labeled')
def avg_area(c_img):
l_img = ndimage.label(c_img > 0)[0]
return np.mean([np.sum(l_img == k) for k in range(1, l_img.max()+1)])
n, _, _ = c_labhist.hist(lab_img[lab_img > 0].ravel(), [
0, 1, 2, 3], rwidth=0.8)
c_labhist.set_title('# Components: %d\nAvg Area: %d' %
(lab_img.max(), avg_area(img_list[i])))
c_lmean.plot(range(i+1),
[avg_area(img_list[k]) for k in range(i+1)], 'r+-')
c_lmean.set_ylim(0, 150)
c_lmean.set_title('Average Area')
# write animation frames
anim_code = FuncAnimation(fig,
update_frame,
frames=len(img_list)-1,
interval=1000,
repeat_delay=2000).to_html5_video()
plt.close('all')
HTML(anim_code)
Distance Map of Real Images¶
We now have a basic idea of how the distance map works we can now try to apply it to real images
%matplotlib inline
from skimage.io import imread
import matplotlib.pyplot as plt
import numpy as np
from scipy.ndimage import distance_transform_edt
bw_img = imread("ext-figures/bonegfiltslice.png")
from skimage.morphology import binary_opening, binary_closing, disk
thresh_img = binary_closing(binary_opening(bw_img < 90, disk(3)), disk(5))
fg_dmap = distance_transform_edt(thresh_img)
bg_dmap = distance_transform_edt(1-thresh_img)
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(20, 6), dpi=100)
ax1.imshow(bw_img, cmap='bone')
ax2.imshow(thresh_img, cmap='bone')
ax3.set_title('Segmentation')
ax3.imshow(fg_dmap, cmap='nipy_spectral')
ax3.set_title('Distance Map\nForeground')
ax4.imshow(bg_dmap, cmap='nipy_spectral')
ax4.set_title('Distance Map\nBackground')
Text(0.5,1,'Distance Map\nBackground')
cortex_img = imread("ext-figures/example_poster.tif")[::2, ::2]/2048
cortex_mask = imread("ext-figures/example_poster_mask.tif")[::1, ::1, 0]/255.0
cortex_dmap = distance_transform_edt(cortex_mask)
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(20, 6), dpi=100)
ax1.imshow(cortex_img, cmap='bone')
ax2.imshow(cortex_mask, cmap='bone')
ax3.imshow(cortex_dmap, cmap='nipy_spectral')
<matplotlib.image.AxesImage at 0x221d3aa9a20>
How can we utilize this information?¶
So we can see in the distance map some information about the size of the object, but the raw values and taking a histogram do not seem to provide this very clearly
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(20, 6), dpi=100)
cortex_dmap_roi = cortex_dmap[25:175, 25:150]
sns.heatmap(cortex_dmap_roi[::5, ::5],
annot=True,
fmt="2.0f",
cmap='nipy_spectral',
ax=ax1,
cbar=True)
ax2.hist(cortex_dmap_roi[cortex_dmap_roi > 0].ravel(), 50);
Thickness Map: What is it?¶
Thickness is a metric for assessing the size and structure of objects in a very generic manner. For every point →x in the image you find the largest sphere which:
- contains that point
- is entirely contained within the object
 Taken from [1]
Taken from [1]
- The image shows a typical object
- The sphere centered at point p with a radius r is the largest that fits
Application¶
- Ideal for spherical and cylindrical objects since it shows their radius
- Also relevant for flow and diffusion since it can highlight bottlenecks in structure
Thickness Map¶
%matplotlib inline
from skimage.io import imread
import matplotlib.pyplot as plt
import numpy as np
from scipy.ndimage import distance_transform_edt
bw_img = imread("ext-figures/bonegfiltslice.png")[::2, ::2]
from skimage.morphology import binary_opening, binary_closing, disk
thresh_img = binary_closing(binary_opening(bw_img < 90, disk(1)), disk(2))
fg_dmap = distance_transform_edt(thresh_img)
bg_dmap = distance_transform_edt(1-thresh_img)
fig, (ax1, ax2, ax3, ax4) = plt.subplots(1, 4, figsize=(20, 6), dpi=100)
ax1.imshow(bw_img, cmap='bone')
ax2.imshow(thresh_img, cmap='bone')
ax3.set_title('Segmentation')
ax3.imshow(fg_dmap, cmap='nipy_spectral')
ax3.set_title('Distance Map\nForeground')
ax4.imshow(bg_dmap, cmap='nipy_spectral')
ax4.set_title('Distance Map\nBackground')
Text(0.5,1,'Distance Map\nBackground')
from tqdm import tqdm
def simple_thickness_func(distance_map):
dm = distance_map.ravel()
th_map = distance_map.ravel().copy()
xx, yy = [v.ravel() for v in np.meshgrid(range(distance_map.shape[1]),
range(distance_map.shape[0]))]
for idx in tqdm(np.argsort(-dm)):
dval = dm[idx]
if dval > 0:
p_mask = (np.square(xx-xx[idx]) +
np.square(yy-yy[idx])) <= np.square(dval)
p_mask &= th_map < dval
th_map[p_mask] = dval
th_map[dm==0] = 0 # make sure zeros stay zero (rounding errors)
return th_map.reshape(distance_map.shape)
fg_th_map = simple_thickness_func(fg_dmap)
bg_th_map = simple_thickness_func(bg_dmap)
100%|█████████████████████████████████████████████████████████████████████████| 21875/21875 [00:00<00:00, 47388.19it/s] 100%|██████████████████████████████████████████████████████████████████████████| 21875/21875 [00:06<00:00, 3354.49it/s]
fig, (ax3, ax4) = plt.subplots(1, 2, figsize=(20, 6), dpi=100)
ax3.imshow(fg_th_map, cmap='nipy_spectral')
ax3.set_title('Thickness Map\nForeground')
ax4.imshow(bg_th_map, cmap='nipy_spectral')
ax4.set_title('Thickness Map\nBackground');
fig, (ax3, ax4) = plt.subplots(1, 2, figsize=(20, 6), dpi=100)
ax3.hist(fg_th_map[fg_th_map>0].ravel(), 20)
ax3.set_title('Thickness Map\nForeground')
ax4.hist(bg_th_map[bg_th_map>0].ravel(), 20)
ax4.set_title('Thickness Map\nBackground');
Distance Maps in 3D¶
Distance maps work in more than two dimensions and can be used in n-d problems although beyond 3D requires serious thought about what the meaning of this distance is.
%matplotlib inline
from skimage.io import imread
import matplotlib.pyplot as plt
import numpy as np
from skimage.util import montage as montage2d
montage_pad = lambda x: montage2d(np.pad(x, [(0,0), (10, 10), (10, 10)], mode = 'constant', constant_values = 0))
from scipy.ndimage import distance_transform_edt
bw_img = 255-imread("../common/data/plateau_border.tif")[:, 100:400, 100:400][:, ::2, ::2]
print('Loaded Image:', bw_img.shape, bw_img.max())
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(18, 8))
ax1.imshow(montage_pad(bw_img[::10]), cmap = 'bone')
ax1.set_title('Axial Slices');
ax2.imshow(montage_pad(bw_img.swapaxes(0,1)[::10]), cmap = 'bone')
ax2.set_title('Sagittal Slices');
Loaded Image: (154, 150, 150) 255
from skimage import measure
from mpl_toolkits.mplot3d.art3d import Poly3DCollection
from tqdm import tqdm_notebook as tqdm
def show_3d_mesh(image, thresholds, edgecolor = 'none', alpha = 0.5):
p = image[::-1].swapaxes(1,2)
cmap = plt.cm.get_cmap('nipy_spectral_r')
fig = plt.figure(figsize=(10, 10))
ax = fig.add_subplot(111, projection='3d')
for i, c_threshold in tqdm(list(enumerate(thresholds))):
verts, faces, _, _ = measure.marching_cubes_lewiner(p, c_threshold)
mesh = Poly3DCollection(verts[faces], alpha=alpha, edgecolor=edgecolor, linewidth = 0.1)
mesh.set_facecolor(cmap(i / len(thresholds))[:3])
ax.add_collection3d(mesh)
ax.set_xlim(0, p.shape[0])
ax.set_ylim(0, p.shape[1])
ax.set_zlim(0, p.shape[2])
ax.view_init(45, 45)
return fig
from skimage.filters import gaussian
smooth_pt_img = gaussian(bw_img[::2, ::2, ::2]*1.0/bw_img.max(), 1.5)
show_3d_mesh(smooth_pt_img, [smooth_pt_img.mean()], alpha = 0.75, edgecolor = 'black');
100%|████████████████████████████████████████████████████████████████████████████████████| 1/1 [00:03<00:00, 3.52s/it]
import plotly.offline as py
import plotly.graph_objs as go
import plotly.figure_factory as FF
py.init_notebook_mode()
verts, faces, _, _ = measure.marching_cubes_lewiner(
smooth_pt_img[:20, :20, :20] , # you can make it bigger but the file-size gets HUUUEGE
smooth_pt_img.mean())
x, y, z = zip(*verts)
ff_fig = FF.create_trisurf(x=x, y=y, z=z,
simplices=faces,
title="Foam Plataeu Border",
aspectratio=dict(x=1, y=1, z=1),
plot_edges=False)
c_mesh = ff_fig['data'][0]
c_mesh.update(lighting=dict(ambient=0.18,
diffuse=1,
fresnel=0.1,
specular=1,
roughness=0.1,
facenormalsepsilon=1e-6,
vertexnormalsepsilon=1e-12))
c_mesh.update(flatshading=False)
py.iplot(ff_fig)
bubble_dist = distance_transform_edt(bw_img)
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(18, 8))
ax1.imshow(montage_pad(bubble_dist[::10]), cmap = 'nipy_spectral')
ax1.set_title('Axial Slices');
ax2.imshow(montage_pad(bubble_dist.swapaxes(0,1)[::10]), cmap = 'nipy_spectral')
ax2.set_title('Sagittal Slices');
Thickness in 3D Images¶
While the examples and demonstrations so far have been shown in 2D, the same exact technique can be applied to 3D data as well. For example for this liquid foam structure

The thickness can be calculated of the background (air) voxels in the same manner.
With care, this can be used as a proxy for bubble size distribution in systems where all the bubbles are connected to it is difficult to identify single ones.

%%time
from skimage.feature import peak_local_max
bubble_candidates = peak_local_max(bubble_dist, min_distance=12)
print('Found', len(bubble_candidates), 'bubbles')
thickness_map = np.zeros(bubble_dist.shape, dtype = np.float32)
xx, yy, zz = np.meshgrid(np.arange(bubble_dist.shape[1]),
np.arange(bubble_dist.shape[0]),
np.arange(bubble_dist.shape[2])
)
# sort candidates by size
sorted_candidates = sorted(bubble_candidates, key = lambda xyz: bubble_dist[tuple(xyz)])
for label_idx, (x,y,z) in enumerate(tqdm(sorted_candidates),1):
cur_bubble_radius = bubble_dist[x,y,z]
cur_bubble = (np.power(xx-float(y),2)+
np.power(yy-float(x),2)+
np.power(zz-float(z),2))<=np.power(cur_bubble_radius,2)
thickness_map[cur_bubble] = cur_bubble_radius
Found 180 bubbles
100%|████████████████████████████████████████████████████████████████████████████████| 180/180 [03:23<00:00, 1.13s/it]
Wall time: 3min 24s
fig, (ax1, ax2, ax3) = plt.subplots(1,3, figsize = (12, 4))
for i, (cax, clabel) in enumerate(zip([ax1, ax2, ax3], ['xy', 'zy', 'zx'])):
cax.imshow(np.max(thickness_map,i).squeeze(), interpolation='none', cmap = 'jet')
cax.set_title('%s Projection' % clabel)
cax.set_xlabel(clabel[0])
cax.set_ylabel(clabel[1])
fig, (ax1, ax2) = plt.subplots(1, 2, figsize=(18, 8))
ax1.imshow(montage_pad(thickness_map[::10]), cmap = 'nipy_spectral')
ax1.set_title('Axial Slices');
ax2.imshow(montage_pad(thickness_map.swapaxes(0,1)[::10]), cmap = 'nipy_spectral')
ax2.set_title('Sagittal Slices');
Interactive 3D Views¶
Here we can show the thickness map in an interactive 3D manner using the ipyvolume tools (probably only works in the Chrome browser)
import ipyvolume as p3
fig = p3.figure()
# create a custom LUT
temp_tf = plt.cm.nipy_spectral(np.linspace(0, 1, 256))
temp_tf[:,3] = np.linspace(-.3, 0.5, 256).clip(0, 1) # make transparency more aggressive
tf = p3.transferfunction.TransferFunction(rgba = temp_tf)
p3.volshow((thickness_map[::2, ::2, ::2]/thickness_map.max()).astype(np.float32),
lighting = True,
max_opacity=0.5,
tf = tf,
controls = True)
p3.show()
p3.save('bubbles.html')
C:\Miniconda3\envs\qbi2018\lib\site-packages\ipyvolume\serialize.py:66: RuntimeWarning: invalid value encountered in true_divide
Failed to display Jupyter Widget of type VBox.
If you're reading this message in the Jupyter Notebook or JupyterLab Notebook, it may mean that the widgets JavaScript is still loading. If this message persists, it likely means that the widgets JavaScript library is either not installed or not enabled. See the Jupyter Widgets Documentation for setup instructions.
If you're reading this message in another frontend (for example, a static rendering on GitHub or NBViewer), it may mean that your frontend doesn't currently support widgets.
C:\Miniconda3\envs\qbi2018\lib\site-packages\ipyvolume\serialize.py:66: RuntimeWarning: invalid value encountered in true_divide
Interfaces / Surfaces¶
Many physical and chemical processes occur at surfaces and interfaces and consequently these structures are important in material science and biology. For this lecture surface and interface will be used interchangebly and refers to the boundary between two different materials (calcified bone and soft tissue, magma and water, liquid and gas) Through segmentation we have identified the unique phases in the sample under investigation.
- Segmentation identifying volumes (3D) or areas (2D)
- Interfaces are one dimension lower corresponding to surface area (3D) or perimeter (2D)
- Interfaces are important for
- connectivity of cell networks, particularly neurons
- material science processes like coarsening or rheological behavior
- chemical processes (surface-bound diffusion, catalyst action)
Surface Area / Perimeter¶
We see that the dilation and erosion affects are strongly related to the surface area of an object: the more surface area the larger the affect of a single dilation or erosion step.
Meshing¶
Constructing a mesh for an image provides very different information than the image data itself. Most crucially this comes when looking at physical processes like deformation.
While the images are helpful for visualizing we rarely have models for quantifying how difficult it is to turn a pixel off
If the image is turned into a mesh we now have a list of vertices and edges. For these vertices and edges we can define forces. For example when looking at stress-strain relationships in mechanics using Hooke's Model →F=k(→x0−→x) the force needed to stretch one of these edges is proportional to how far it is stretched.
Marching Cubes¶
Why¶
Voxels are very poor approximations for the surface and are very rough (they are either normal to the x, y, or z axis and nothing between). Because of their inherently orthogonal surface normals, any analysis which utilizes the surface normal to calculate another value (growth, curvature, etc) is going to be very inaccurate at best and very wrong at worst.
How¶
The image is processed one voxel at a time and the neighborhood (not quite the same is the morphological definition) is checked at every voxel. From this configuration of values, faces are added to the mesh to incorporate the most simple surface which would explain the values.
Marching tetrahedra is for some applications a better suited approach
Curvature¶
Curvature is a metric related to the surface or interface between phases or objects.
- It is most easily understood in its 1D sense or being the radius of the circle that matchs the local shape of a curve
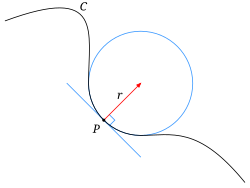
- Mathematically it is defined as
- Thus a low curvature means the value means a very large radius and high curvature means a very small radius
Curvature: Surface Normal¶
In order to meaningfully talk about curvatures of surfaces, we first need to define a consistent frame of reference for examining the surface of an object. We thus define a surface normal vector as a vector oriented orthogonally to the surface away from the interior of the object →→N


Curvature: 3D¶
With the notion of surface normal defined (→N), we can define many curvatures at point →x on the surface.
- This is because there are infinitely many planes which contain both point →x and →N
- More generally we can define an angle θ about which a single plane containing both can be freely rotated
- We can then define two principal curvatures by taking the maximum and minimum of this curve
Mean Curvature¶
The mean of the two principal curvatures H=12(κ1+κ2)
Gaussian Curvature¶
The mean of the two principal curvatures K=κ1κ2
- positive for spheres (or spherical inclusions)
- curvatures agree in sign
- negative for saddles (hyperboloid surfaces)
- curvatures disgree in sign
- 0 for planes
Curvature: 3D Examples¶
Examining a complex structure with no meaningful ellipsoidal or watershed model. The images themselves show the type of substructures and shapes which are present in the sample.

- gaussian curvature: the comparative amount of surface at, above, and below 0
- from spherical particles into annealed mesh of balls

Characteristic Shape¶
Characteristic shape can be calculated by measuring principal curvatures and normalizing them by scaling to the structure size. A distribution of these curvatures then provides shape information on a structure indepedent of the size.
For example a structure transitioning from a collection of perfectly spherical particles to a annealed solid will go from having many round spherical faces with positive gaussian curvature to many saddles and more complicated structures with 0 or negative curvature.
Curvature: Take Home Message¶
It provides another metric for characterizing complex shapes
- Particularly useful for examining interfaces
- Folds, saddles, and many other types of points are not characterized well be ellipsoids or thickness maps
- Provides a scale-free metric for assessing structures
- Can provide visual indications of structural changes
Other Techniques¶
There are hundreds of other techniques which can be applied to these complicated structures, but they go beyond the scope of this course. Many of them are model-based which means they work well but only for particular types of samples or images. Of the more general techniques several which are easily testable inside of FIJI are
- Directional Analysis = Looking at the orientation of different components using Fourier analysis (Analyze → Directionality)
- Tubeness / Surfaceness (Plugins → Analyze →) characterize binary images and the shape at each point similar to curvature but with a different underlying model
- Fractal Dimensionality = A metric for assessing the structure as you scale up and down by examining various spatial relationships
- Ma, D., Stoica, A. D., & Wang, X.-L. (2009). Power-law scaling and fractal nature of medium-range order in metallic glasses. Nature Materials, 8(1), 30–4. doi:10.1038/nmat2340
- Two (or more) point correlation functions = Used in theoretical material science and physics to describe random materials and can be used to characterize distances, orientations, and organization in complex samples
- Jiao, Y., Stillinger, F., & Torquato, S. (2007). Modeling heterogeneous materials via two-point correlation functions: Basic principles. Physical Review E, 76(3). doi:10.1103/PhysRevE.76.031110
- Andrey, P., Kiêu, K., Kress, C., Lehmann, G., Tirichine, L., Liu, Z., … Debey, P. (2010). Statistical analysis of 3D images detects regular spatial distributions of centromeres and chromocenters in animal and plant nuclei. PLoS Computational Biology, 6(7), e1000853. doi:10.1371/journal.pcbi.1000853
- Haghpanahi, M., & Miramini, S. (2008). Extraction of morphological parameters of tissue engineering scaffolds using two-point correlation function, 463–466. Retrieved from http://portal.acm.org/citation.cfm?id=1713360.1713456
Sample Textures¶
{r}
text_imgs<-expand.grid(x=1:16,y=1:16) %>%
mutate(val_x=x%%2,
val_y=y%%2,
val_xy=(x%%2+y%%2)/2.0,
val_x2y=(x%%4+y%%2)/2.5,
val_x2y2=(x%%4+y%%3)/3.5,
val_xya=(x+y)%%3/2.0) %>%
melt(id.vars=c("x","y"))
text_imgs %>% ggplot(aes(x,y))+
geom_raster(aes(fill=value))+
facet_wrap(~variable)+
coord_equal()+
theme_bw(10)
{r}
text_imgs %>% ggplot(aes(value))+
geom_density(aes(fill=variable),alpha=0.5)+
theme_bw(10)
Co-occurrence matrix¶
CΔx,Δy(i,j)=n∑x=1m∑y=1{1,if I(x,y)=i and I(x+Δx,y+Δy)=j0,otherwise- As text → for every possible offset →r = (Δx, Δy) given the current intensity (i) how likely is it that value at a point offset by that amount is j.
- This is similar to the two point correlation function but is a 4D representation instead of just a 2D
- In order to use this we need to discretize the image into bins
{r}
bin_it<-function(x) as.numeric(as.factor(cut_interval(x,4)))
bin_image<-text_imgs %>%
mutate(value=bin_it(value))
bin_image %>%
group_by(variable,value) %>%
summarize(count=n()) %>%
ggplot(aes(value,count))+
geom_bar(aes(fill=variable),alpha=0.5,stat="identity",pos="dodge")+
facet_grid(~variable)+
theme_bw(10)
Calculating the Matrix¶
{r}
bin_image %>%
merge(bin_image,by=c("variable")) %>%
mutate(dx=x.y-x.x,dy=y.y-y.x) %>%
subset((abs(dx)<4) & (abs(dy)<4)) %>%
group_by(variable,dx,dy,value.x,value.y) %>%
summarize(count=n()) %>%
mutate(ij=paste0(value.x,"->",value.y)) %>%
ggplot(aes(dx,dy))+
geom_raster(aes(fill=count))+
facet_grid(variable~ij)+
theme_bw(10)
Overlapping¶
{r}
bin_image %>%
merge(bin_image,by=c("variable")) %>%
mutate(dx=x.y-x.x,dy=y.y-y.x) %>%
subset((abs(dx)<5) & (abs(dy)<5)) %>%
group_by(variable,dx,dy,value.x,value.y) %>%
summarize(count=n()) %>%
mutate(ij=paste0(value.x,"->",value.y)) %>%
ggplot(aes(dx,dy))+
geom_contour(aes(z=count,color=ij))+
facet_wrap(~variable)+
theme_bw(10)
Simple Correlation¶
Using the mean difference (E[i−j]) instead of all of the i,j pair possiblities
{r}
bin_image %>%
merge(bin_image,by=c("variable")) %>%
mutate(dx=x.y-x.x,dy=y.y-y.x,dv=value.y-value.x) %>%
subset((abs(dx)<4) & (abs(dy)<4)) %>%
group_by(variable,dx,dy,value.x,value.y) %>%
summarize(score=mean(dv)) %>%
ggplot(aes(dx,dy))+
geom_raster(aes(fill=score))+
facet_wrap(~variable)+
coord_equal()+
theme_bw(10)
Radial Coordinates¶
Rather than using Δx and Δy we can also show the relative positions
{r}
bin_image %>%
merge(bin_image,by=c("variable")) %>%
mutate(dr=sqrt((x.y-x.x)**2+(y.y-y.x)**2),
dtheta=180/pi*atan((y.y-y.x)/(x.y-x.x)),
dv=value.y-value.x) %>%
subset(dr<6 & !is.na(dtheta)) %>%
mutate(dr=cut_interval(dr,10),
dtheta=cut_interval(dtheta,8)) %>%
group_by(variable,dr,dtheta,value.x,value.y) %>%
summarize(score=mean(dv)) %>%
ggplot(aes(dtheta,dr))+
geom_tile(aes(fill=score))+
facet_wrap(~variable)+
coord_polar(theta="x")+
scale_fill_distiller(palette = "Spectral")+
theme_bw(10)
Applying Texture to Brain¶
%matplotlib inline
from skimage.io import imread
import matplotlib.pyplot as plt
import numpy as np
from skimage.color import label2rgb
import seaborn as sns
cortex_img = np.clip(imread("ext-figures/example_poster.tif")
[::2, ::2]/270, 0, 255).astype(np.uint8)
cortex_mask = imread("ext-figures/example_poster_mask.tif")[::1, ::1, 0]/255.0
# show the slice and threshold
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize=(11, 5))
ax1.imshow(cortex_img, cmap='gray')
ax1.axis('off')
ax1.set_title('Image')
ax2.imshow(cortex_mask, cmap='gray')
ax2.axis('off')
ax2.set_title('Segmentation')
# here we mark the threshold on the original image
ax3.imshow(label2rgb(cortex_mask > 0, cortex_img, bg_label=0))
ax3.axis('off')
ax3.set_title('Overlayed');
Tiling¶
Here we divide the image up into unique tiles for further processing
xx, yy = np.meshgrid(
np.arange(cortex_img.shape[1]),
np.arange(cortex_img.shape[0]))
region_labels = (xx//48) * 64+yy//48
region_labels = region_labels.astype(int)
sns.heatmap(region_labels[::48, ::48].astype(int),
annot=True,
fmt="03d",
cmap='nipy_spectral',
cbar=False,
);
Calculating Texture¶
Here we calculate the texture by using a tool called the gray level co-occurrence matrix which are part of the features library in skimage. We focus on two metrics in this, specifically dissimilarity and correlation which we calculate for each tile. We then want to see which of these parameters correlated best with belonging to a nerve fiber.
# compute some GLCM properties each patch
from skimage.feature import greycomatrix, greycoprops
from tqdm import tqdm_notebook as tqdm
grayco_prop_list = ['contrast', 'dissimilarity',
'homogeneity', 'energy',
'correlation', 'ASM']
prop_imgs = {}
for c_prop in grayco_prop_list:
prop_imgs[c_prop] = np.zeros_like(cortex_img, dtype=np.float32)
score_img = np.zeros_like(cortex_img, dtype=np.float32)
out_df_list = []
for patch_idx in tqdm(np.unique(region_labels)):
xx_box, yy_box = np.where(region_labels == patch_idx)
glcm = greycomatrix(cortex_img[xx_box.min():xx_box.max(),
yy_box.min():yy_box.max()],
[5], [0], 256, symmetric=True, normed=True)
mean_score = np.mean(cortex_mask[region_labels == patch_idx])
score_img[region_labels == patch_idx] = mean_score
out_row = dict(
intensity_mean=np.mean(cortex_img[region_labels == patch_idx]),
intensity_std=np.std(cortex_img[region_labels == patch_idx]),
score=mean_score)
for c_prop in grayco_prop_list:
out_row[c_prop] = greycoprops(glcm, c_prop)[0, 0]
prop_imgs[c_prop][region_labels == patch_idx] = out_row[c_prop]
out_df_list += [out_row]
Failed to display Jupyter Widget of type HBox.
If you're reading this message in the Jupyter Notebook or JupyterLab Notebook, it may mean that the widgets JavaScript is still loading. If this message persists, it likely means that the widgets JavaScript library is either not installed or not enabled. See the Jupyter Widgets Documentation for setup instructions.
If you're reading this message in another frontend (for example, a static rendering on GitHub or NBViewer), it may mean that your frontend doesn't currently support widgets.
C:\Miniconda3\envs\qbi2018\lib\site-packages\skimage\feature\texture.py:109: FutureWarning: Conversion of the second argument of issubdtype from `float` to `np.floating` is deprecated. In future, it will be treated as `np.float64 == np.dtype(float).type`. if np.issubdtype(image.dtype, np.float):
# show the slice and threshold
fig, m_axs = plt.subplots(2, 4, figsize=(20, 10))
n_axs = m_axs.flatten()
ax1 = n_axs[0]
ax2 = n_axs[1]
ax1.imshow(cortex_img, cmap='gray')
ax1.axis('off')
ax1.set_title('Image')
ax2.imshow(score_img, cmap='gray')
ax2.axis('off')
ax2.set_title('Regions')
for c_ax, c_prop in zip(n_axs[2:], grayco_prop_list):
c_ax.imshow(prop_imgs[c_prop], cmap='gray')
c_ax.axis('off')
c_ax.set_title('{} Image'.format(c_prop))
import pandas as pd
out_df = pd.DataFrame(out_df_list)
out_df['positive_score'] = out_df['score'].map(
lambda x: 'FG' if x > 0 else 'BG')
out_df.describe()
| ASM | contrast | correlation | dissimilarity | energy | homogeneity | intensity_mean | intensity_std | score | |
|---|---|---|---|---|---|---|---|---|---|
| count | 99.000000 | 99.000000 | 99.000000 | 99.000000 | 99.000000 | 99.000000 | 99.000000 | 99.000000 | 99.000000 |
| mean | 0.045523 | 888.349768 | 0.468967 | 15.933864 | 0.128972 | 0.179556 | 61.214533 | 28.922878 | 0.159290 |
| std | 0.096791 | 1736.226189 | 0.268096 | 10.016607 | 0.170833 | 0.173307 | 32.747924 | 20.736974 | 0.310034 |
| min | 0.000366 | 14.721364 | -0.094922 | 1.666023 | 0.019123 | 0.033100 | 1.274123 | 3.241296 | 0.000000 |
| 25% | 0.000626 | 249.179078 | 0.247538 | 11.847264 | 0.025010 | 0.070740 | 39.180681 | 14.174057 | 0.000000 |
| 50% | 0.000904 | 334.321682 | 0.504085 | 13.734549 | 0.030073 | 0.082297 | 62.908854 | 20.017975 | 0.000000 |
| 75% | 0.036697 | 779.776342 | 0.677313 | 18.028875 | 0.191400 | 0.250455 | 80.434896 | 35.653407 | 0.078170 |
| max | 0.424854 | 13619.651391 | 0.958805 | 82.260229 | 0.651809 | 0.718922 | 131.299479 | 94.529072 | 1.000000 |
import seaborn as sns
sns.pairplot(out_df,
hue='positive_score',
kind="reg")
<seaborn.axisgrid.PairGrid at 0x26eaca12b38>