#!/usr/bin/env python
# coding: utf-8
# Patrick BROCKMANN
# Software engineer at LSCE (Climate and Environment Sciences Laboratory)
# 
#
# Date: 18 November 2015
# Release: 0.7
# ## Proposal for a structured spreadsheet for Paleo Data to interface with the Linked Paleo Data container
# The Linked Paleo Data (LiPD) container based on the [Linked Data JSON (JSON-LD) format](https://www.authorea.com/users/17200/articles/19163/_show_article) is a practical solution to the problem of organizing and storing hierarchical paleoclimate data in a generalizable schema. This is an important step forward towards standardizing the representation and linkage of diverse paleoclimate datasets.
#
# In this IPython notebook, I have experimental converters to interact with the LiPD container using ordinary spreadsheets. The motivation to create this method is guided by the fact that the paleoclimate community uses mainly spreadsheets to edit and store the data and the metadata of their measurements, and not JSON-based formats. What is missing is a way to convert such spreadsheet-based data to LiPD format and vice versa.
#
# Working directly with LiPD has two other disadvantages:
# 1. JSON notation will never be as easy to edit and modify than a spreadsheet document;
# 1. The LiPD container refers to a headerless CSV data file which requires the user to continually navigate the nested attributes of the LiPD file in order to figure out which parameters correspond to which columns in the CSV file.
#
# Therefore, I propose to stick with the use of spreadsheets but standardize them into a structured spreadsheet where the data and the metadata are stored in two separate worksheets of the same spreadsheet document. The [dot notation](https://developer.mozilla.org/en-US/docs/Web/JavaScript/Reference/Operators/Property_Accessors) is used to represent the hierarchical nature of the metadata attributes. Following the nomenclature of LiPD, I call this structured spreadsheet **PDS** for **Paleo Data Spreadsheet**.
#
# With a PDS, users can directly edit their data in an ordinary spreadsheet program like Excel or OpenOffice and later convert them to LiPD, which is a good container for storing data in a document database like mongoDB (since it uses JSON).
# In addition, I have implemented converters to transform PDS to [python pandas dataframes](http://pandas.pydata.org/pandas-docs/version/0.17.0/dsintro.html#dataframe), which are convenient for subsequent data analysis in e.g. an IPython notebook.
# ### The Paleo Data Spreadsheet (PDS) structure
# - 2 worksheets: Data and Metadata.
# - The Data worksheet presents the data in a matrix with named columns for each parameter and rows for each sample, as in a CSV file.
# - The Metadata worksheet has 2 columns corresponding to the Attribute and Value of all the parameters described in the Data worksheet. There are no headers. Hierarchical attributes are denoted by the dot notation (e.g. parameter.attribute) in a Attribute column and their values are contained in a Value column. If there is no corresponding parameter in the Data worksheet, then it is assumed that the attribute is global (e.g. filename).
# - Missing data are left as empty cells.
# - No comments are allowed in the Data and Metadata worksheets but other worksheets can be added for this purpose.
#
# The Data worksheet: 
# The Metadata worksheet: 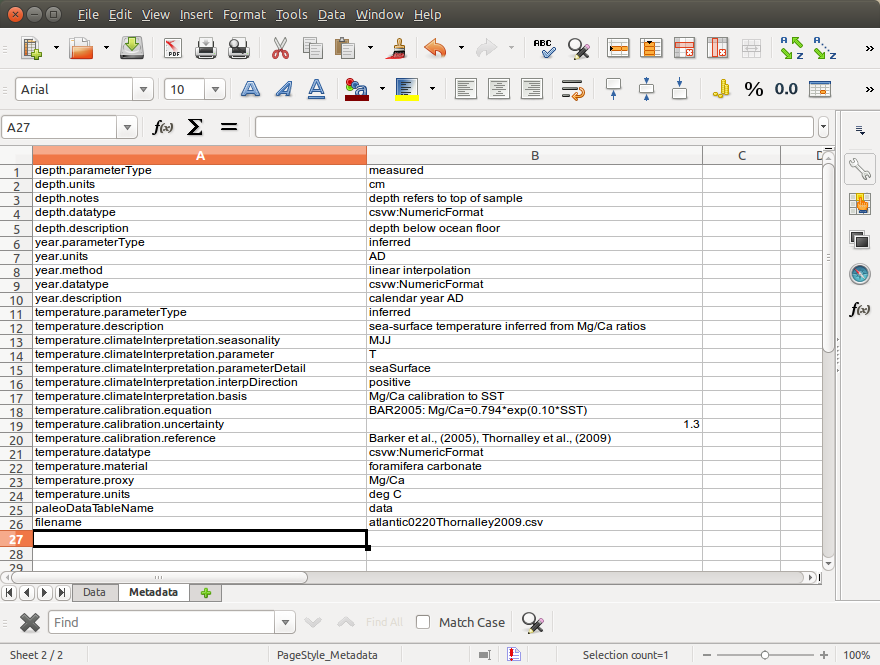
# Notes:
# 1. Currently, pandas does not yet support the reading and the writing of ODS (Open Document Spreadsheets) but there are many requests for this feature and it should be feasible soon ([pandas/issue 2311](https://github.com/pydata/pandas/issues/2311)).
#
# 1. The LiPD container refers to a headerless CSV file where the data are stored. Each column is therefore referenced only by a column number and is very poorly documented, and this could lead to confusion. I think it would be safer and clearer to use the first row as a header to name the columns by the parameter names.
#
# 1. The LiPD container can contain a list object for values, e.g. [{ "author": [{"name" : "N1"}, {"name" : "N2"}, {"name" : "N3"}] }]. It would be simpler to disable this possibility and have only unique values.
#
# 1. The spreadsheet cells must be formatted correctly, i.e. numbers cells are specified as number and not text. Same for dates.
#
# 1. A compliance checker needs to be built to check that input files conform to the PDS structure, something like the [netCDF CF-checker](http://cfconventions.org/compliance-checker.html).
#
# 1. The working group of [PAGES (Past Global Changes)](http://www.pages-igbp.org/) called [2K Network](http://www.pages-igbp.org/ini/wg/2k-network/data) proposes to collect data using a [PAGES2k/NOAA metadata template](http://www.pages-igbp.org/download/docs/working_groups/2k_network/2014-2kphase2-noaa-wdc-paleo-template.xlsx).
# This template is very detailled but not easilly convertible to other structures such a pandas dataframes or LiPD container.
#
# 1. A RESTful web service could be implemented to visualize a PDS file as an HTML page with interactive plots. [Bokeh](http://bokeh.pydata.org/en/latest/), a python interactive visualization library, would be useful for this purpose.
# ### Converting from one structure to another with PDSlib
# PDSlib is a python module that offers two-way converters for different structures: PDS, pandas dataframes (df), and LiPD container.
#
#  #
# - **aLiPD = PDS_to_LiPD (PDS_inputfile, verbose=True)**
# * Convert a PDS to a LiPD container
# * If verbose is True, warning messages will be printed
# - **LiPD_to_PDS (aLiPD, PDS_outputfile)**
# * Convert a LiPD container to a PDS
# - **dfD, dfM = PDS_to_df (PDS_inputfile)**
# * Convert a PDS to 2 pandas dataframes for Data and Metadata, respectively
# - **df_to_PDS (dfD, dfM, PDS_outputfile)**
# * Convert 2 pandas dataframes for Data and Metadata to a PDS
# - **aLiPD = df_to_LiPD (dfD, dfM, verbose=True)**
# * Convert 2 pandas dataframes for Data and Metadata to a LiPD container
# * If verbose is True, warning messages will be printed
# - **dfD, dfM = LiPD_to_df (aLiPD)**
# * Convert a LiPD container to 2 pandas dataframes for Data and Metadata, respectively
# - **df_plot (dfD, xCol, [yCols], width=600, height=600)**
# * Convert a data pandas dataframe to an interactive plot using xCol and yCols, respectively as xaxis and yaxis
# * The function relies on bokeh module
# - **PDS_to_html (PDS_inputfile, xCol, [yCols], HTML_outputfile, title=None, width=600, height=600)**
# * Convert a PDS to an html file with Data as an interactive plot and Metadata as a table
# * The function relies on bokeh module
# ### Convert a PDS to pandas dataframes
# In[1]:
import PDSlib
# In[2]:
dfD, dfM = PDSlib.PDS_to_df('PDS_01.xls')
# In[3]:
dfD
# In[4]:
dfM
# ### Convert the pandas dataframes to a LiPD container
# In[5]:
a_LiPD = PDSlib.df_to_LiPD(dfD, dfM, verbose=True)
import json
print json.dumps(a_LiPD, sort_keys=False, indent=4, separators=(',', ': '))
# This exemple comes from the article [Linked Data JSON (JSON-LD) format](https://www.authorea.com/users/17200/articles/19163/_show_article) but somes keys have been changed from column numbers to parameter names accordingly the note #2.
# ### Convert a LiPD container to pandas dataframes
# In[6]:
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
# In[7]:
dfD
# In[8]:
dfM
# ### Convert pandas dataframes to a PDS
# In[9]:
PDSlib.df_to_PDS(dfD, dfM, 'PDS_02.xls')
# PDS_01.xls is identical to PDS_02.xls proof that conversions are correct.
#
# PDS ===> LiPD ===> PDS
# ### Direct conversion from a PDS to a LiPD container
# In[10]:
a = PDSlib.PDS_to_LiPD('PDS_Uemura.xls', verbose=True)
import json
print json.dumps(a, sort_keys=False, indent=4, separators=(',', ': '))
# ### Direct convertion from a LiPD container to a PDS
# In[11]:
PDSlib.LiPD_to_PDS(a, 'PDS_04.xls')
# ### Convert the data pandas dataframe as an interactive plot with [bokeh]( http://bokeh.pydata.org/en/latest/)
# In[12]:
dfD, dfM = PDSlib.PDS_to_df('PDS_Uemura.xls')
# In[13]:
dfM.sort_values(by='Attribute')
# In[14]:
dfD.columns
# In[15]:
PDSlib.df_plot(dfD, 1, [2,3,4])
# ### Convert a PDS to an html file with Data as an interactive plot and Metadata as a table with bokeh
# In[16]:
PDSlib.PDS_to_html("PDS_Uemura.xls", 1, [2,3,4], "PDS_Uemura.html", width=600, height=400)
# [Open the html file created](PDS_Uemura.html)
# ## Below are some tests to deal with values entered as lists
# ### A conversion test from a BibJSON that describes publication metadata
# In[17]:
a_LiPD = [{
"author": [
{"name" : "Thornalley, D.J.R"},
{"name" : "Elderfield, H."},
{"name" : "McCave, N"}
],
"type" : "article",
"identifier" :
{"type": "doi",
"id": "10.1038/nature07717",
"url": "http://dx.doi.org/10.1038/nature07717"}
,
"pubYear": 2009
}]
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
dfM
# ### A conversion test with a GeoJSON
# In[18]:
a_LiPD = { "type": "FeatureCollection",
"features": [
{ "type": "Feature",
"geometry": {"type": "Point", "coordinates": [102.0, 0.5]},
"properties": {"prop0": "value0"}
},
{ "type": "Feature",
"geometry": {
"type": "LineString",
"coordinates": [
[102.0, 0.0], [103.0, 1.0], [104.0, 0.0], [105.0, 1.0]
]
},
"properties": {
"prop0": "value0",
"prop1": 0.0
}
},
{ "type": "Feature",
"geometry": {
"type": "Polygon",
"coordinates": [
[ [100.0, 0.0], [101.0, 0.0], [101.0, 1.0],
[100.0, 1.0], [100.0, 0.0] ]
]
},
"properties": {
"prop0": "value0",
"prop1": {"this": "that"}
}
}
]
}
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
dfM
# These last 2 conversions examples are not bidirectional since list items in LiPD containers are transformed using numbers for each item when PDSlib.LiPD_to_df is used:
# - author[] to author.1, author.2, author.3
# - features[] to features.1, features.2
#
# Should lists be allowed in LiPD ? If so, more work is required to recreate a list from numbered items when using PDSlib.df_to_LiPD
# - author.1, author.2, author.3 to author[]
# - features.1, features.2 to features[]
# In[ ]:
# In[ ]:
#
# - **aLiPD = PDS_to_LiPD (PDS_inputfile, verbose=True)**
# * Convert a PDS to a LiPD container
# * If verbose is True, warning messages will be printed
# - **LiPD_to_PDS (aLiPD, PDS_outputfile)**
# * Convert a LiPD container to a PDS
# - **dfD, dfM = PDS_to_df (PDS_inputfile)**
# * Convert a PDS to 2 pandas dataframes for Data and Metadata, respectively
# - **df_to_PDS (dfD, dfM, PDS_outputfile)**
# * Convert 2 pandas dataframes for Data and Metadata to a PDS
# - **aLiPD = df_to_LiPD (dfD, dfM, verbose=True)**
# * Convert 2 pandas dataframes for Data and Metadata to a LiPD container
# * If verbose is True, warning messages will be printed
# - **dfD, dfM = LiPD_to_df (aLiPD)**
# * Convert a LiPD container to 2 pandas dataframes for Data and Metadata, respectively
# - **df_plot (dfD, xCol, [yCols], width=600, height=600)**
# * Convert a data pandas dataframe to an interactive plot using xCol and yCols, respectively as xaxis and yaxis
# * The function relies on bokeh module
# - **PDS_to_html (PDS_inputfile, xCol, [yCols], HTML_outputfile, title=None, width=600, height=600)**
# * Convert a PDS to an html file with Data as an interactive plot and Metadata as a table
# * The function relies on bokeh module
# ### Convert a PDS to pandas dataframes
# In[1]:
import PDSlib
# In[2]:
dfD, dfM = PDSlib.PDS_to_df('PDS_01.xls')
# In[3]:
dfD
# In[4]:
dfM
# ### Convert the pandas dataframes to a LiPD container
# In[5]:
a_LiPD = PDSlib.df_to_LiPD(dfD, dfM, verbose=True)
import json
print json.dumps(a_LiPD, sort_keys=False, indent=4, separators=(',', ': '))
# This exemple comes from the article [Linked Data JSON (JSON-LD) format](https://www.authorea.com/users/17200/articles/19163/_show_article) but somes keys have been changed from column numbers to parameter names accordingly the note #2.
# ### Convert a LiPD container to pandas dataframes
# In[6]:
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
# In[7]:
dfD
# In[8]:
dfM
# ### Convert pandas dataframes to a PDS
# In[9]:
PDSlib.df_to_PDS(dfD, dfM, 'PDS_02.xls')
# PDS_01.xls is identical to PDS_02.xls proof that conversions are correct.
#
# PDS ===> LiPD ===> PDS
# ### Direct conversion from a PDS to a LiPD container
# In[10]:
a = PDSlib.PDS_to_LiPD('PDS_Uemura.xls', verbose=True)
import json
print json.dumps(a, sort_keys=False, indent=4, separators=(',', ': '))
# ### Direct convertion from a LiPD container to a PDS
# In[11]:
PDSlib.LiPD_to_PDS(a, 'PDS_04.xls')
# ### Convert the data pandas dataframe as an interactive plot with [bokeh]( http://bokeh.pydata.org/en/latest/)
# In[12]:
dfD, dfM = PDSlib.PDS_to_df('PDS_Uemura.xls')
# In[13]:
dfM.sort_values(by='Attribute')
# In[14]:
dfD.columns
# In[15]:
PDSlib.df_plot(dfD, 1, [2,3,4])
# ### Convert a PDS to an html file with Data as an interactive plot and Metadata as a table with bokeh
# In[16]:
PDSlib.PDS_to_html("PDS_Uemura.xls", 1, [2,3,4], "PDS_Uemura.html", width=600, height=400)
# [Open the html file created](PDS_Uemura.html)
# ## Below are some tests to deal with values entered as lists
# ### A conversion test from a BibJSON that describes publication metadata
# In[17]:
a_LiPD = [{
"author": [
{"name" : "Thornalley, D.J.R"},
{"name" : "Elderfield, H."},
{"name" : "McCave, N"}
],
"type" : "article",
"identifier" :
{"type": "doi",
"id": "10.1038/nature07717",
"url": "http://dx.doi.org/10.1038/nature07717"}
,
"pubYear": 2009
}]
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
dfM
# ### A conversion test with a GeoJSON
# In[18]:
a_LiPD = { "type": "FeatureCollection",
"features": [
{ "type": "Feature",
"geometry": {"type": "Point", "coordinates": [102.0, 0.5]},
"properties": {"prop0": "value0"}
},
{ "type": "Feature",
"geometry": {
"type": "LineString",
"coordinates": [
[102.0, 0.0], [103.0, 1.0], [104.0, 0.0], [105.0, 1.0]
]
},
"properties": {
"prop0": "value0",
"prop1": 0.0
}
},
{ "type": "Feature",
"geometry": {
"type": "Polygon",
"coordinates": [
[ [100.0, 0.0], [101.0, 0.0], [101.0, 1.0],
[100.0, 1.0], [100.0, 0.0] ]
]
},
"properties": {
"prop0": "value0",
"prop1": {"this": "that"}
}
}
]
}
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
dfM
# These last 2 conversions examples are not bidirectional since list items in LiPD containers are transformed using numbers for each item when PDSlib.LiPD_to_df is used:
# - author[] to author.1, author.2, author.3
# - features[] to features.1, features.2
#
# Should lists be allowed in LiPD ? If so, more work is required to recreate a list from numbered items when using PDSlib.df_to_LiPD
# - author.1, author.2, author.3 to author[]
# - features.1, features.2 to features[]
# In[ ]:
# In[ ]:



 #
# - **aLiPD = PDS_to_LiPD (PDS_inputfile, verbose=True)**
# * Convert a PDS to a LiPD container
# * If verbose is True, warning messages will be printed
# - **LiPD_to_PDS (aLiPD, PDS_outputfile)**
# * Convert a LiPD container to a PDS
# - **dfD, dfM = PDS_to_df (PDS_inputfile)**
# * Convert a PDS to 2 pandas dataframes for Data and Metadata, respectively
# - **df_to_PDS (dfD, dfM, PDS_outputfile)**
# * Convert 2 pandas dataframes for Data and Metadata to a PDS
# - **aLiPD = df_to_LiPD (dfD, dfM, verbose=True)**
# * Convert 2 pandas dataframes for Data and Metadata to a LiPD container
# * If verbose is True, warning messages will be printed
# - **dfD, dfM = LiPD_to_df (aLiPD)**
# * Convert a LiPD container to 2 pandas dataframes for Data and Metadata, respectively
# - **df_plot (dfD, xCol, [yCols], width=600, height=600)**
# * Convert a data pandas dataframe to an interactive plot using xCol and yCols, respectively as xaxis and yaxis
# * The function relies on bokeh module
# - **PDS_to_html (PDS_inputfile, xCol, [yCols], HTML_outputfile, title=None, width=600, height=600)**
# * Convert a PDS to an html file with Data as an interactive plot and Metadata as a table
# * The function relies on bokeh module
# ### Convert a PDS to pandas dataframes
# In[1]:
import PDSlib
# In[2]:
dfD, dfM = PDSlib.PDS_to_df('PDS_01.xls')
# In[3]:
dfD
# In[4]:
dfM
# ### Convert the pandas dataframes to a LiPD container
# In[5]:
a_LiPD = PDSlib.df_to_LiPD(dfD, dfM, verbose=True)
import json
print json.dumps(a_LiPD, sort_keys=False, indent=4, separators=(',', ': '))
# This exemple comes from the article [Linked Data JSON (JSON-LD) format](https://www.authorea.com/users/17200/articles/19163/_show_article) but somes keys have been changed from column numbers to parameter names accordingly the note #2.
# ### Convert a LiPD container to pandas dataframes
# In[6]:
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
# In[7]:
dfD
# In[8]:
dfM
# ### Convert pandas dataframes to a PDS
# In[9]:
PDSlib.df_to_PDS(dfD, dfM, 'PDS_02.xls')
# PDS_01.xls is identical to PDS_02.xls proof that conversions are correct.
#
# PDS ===> LiPD ===> PDS
# ### Direct conversion from a PDS to a LiPD container
# In[10]:
a = PDSlib.PDS_to_LiPD('PDS_Uemura.xls', verbose=True)
import json
print json.dumps(a, sort_keys=False, indent=4, separators=(',', ': '))
# ### Direct convertion from a LiPD container to a PDS
# In[11]:
PDSlib.LiPD_to_PDS(a, 'PDS_04.xls')
# ### Convert the data pandas dataframe as an interactive plot with [bokeh]( http://bokeh.pydata.org/en/latest/)
# In[12]:
dfD, dfM = PDSlib.PDS_to_df('PDS_Uemura.xls')
# In[13]:
dfM.sort_values(by='Attribute')
# In[14]:
dfD.columns
# In[15]:
PDSlib.df_plot(dfD, 1, [2,3,4])
# ### Convert a PDS to an html file with Data as an interactive plot and Metadata as a table with bokeh
# In[16]:
PDSlib.PDS_to_html("PDS_Uemura.xls", 1, [2,3,4], "PDS_Uemura.html", width=600, height=400)
# [Open the html file created](PDS_Uemura.html)
# ## Below are some tests to deal with values entered as lists
# ### A conversion test from a BibJSON that describes publication metadata
# In[17]:
a_LiPD = [{
"author": [
{"name" : "Thornalley, D.J.R"},
{"name" : "Elderfield, H."},
{"name" : "McCave, N"}
],
"type" : "article",
"identifier" :
{"type": "doi",
"id": "10.1038/nature07717",
"url": "http://dx.doi.org/10.1038/nature07717"}
,
"pubYear": 2009
}]
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
dfM
# ### A conversion test with a GeoJSON
# In[18]:
a_LiPD = { "type": "FeatureCollection",
"features": [
{ "type": "Feature",
"geometry": {"type": "Point", "coordinates": [102.0, 0.5]},
"properties": {"prop0": "value0"}
},
{ "type": "Feature",
"geometry": {
"type": "LineString",
"coordinates": [
[102.0, 0.0], [103.0, 1.0], [104.0, 0.0], [105.0, 1.0]
]
},
"properties": {
"prop0": "value0",
"prop1": 0.0
}
},
{ "type": "Feature",
"geometry": {
"type": "Polygon",
"coordinates": [
[ [100.0, 0.0], [101.0, 0.0], [101.0, 1.0],
[100.0, 1.0], [100.0, 0.0] ]
]
},
"properties": {
"prop0": "value0",
"prop1": {"this": "that"}
}
}
]
}
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
dfM
# These last 2 conversions examples are not bidirectional since list items in LiPD containers are transformed using numbers for each item when PDSlib.LiPD_to_df is used:
# - author[] to author.1, author.2, author.3
# - features[] to features.1, features.2
#
# Should lists be allowed in LiPD ? If so, more work is required to recreate a list from numbered items when using PDSlib.df_to_LiPD
# - author.1, author.2, author.3 to author[]
# - features.1, features.2 to features[]
# In[ ]:
# In[ ]:
#
# - **aLiPD = PDS_to_LiPD (PDS_inputfile, verbose=True)**
# * Convert a PDS to a LiPD container
# * If verbose is True, warning messages will be printed
# - **LiPD_to_PDS (aLiPD, PDS_outputfile)**
# * Convert a LiPD container to a PDS
# - **dfD, dfM = PDS_to_df (PDS_inputfile)**
# * Convert a PDS to 2 pandas dataframes for Data and Metadata, respectively
# - **df_to_PDS (dfD, dfM, PDS_outputfile)**
# * Convert 2 pandas dataframes for Data and Metadata to a PDS
# - **aLiPD = df_to_LiPD (dfD, dfM, verbose=True)**
# * Convert 2 pandas dataframes for Data and Metadata to a LiPD container
# * If verbose is True, warning messages will be printed
# - **dfD, dfM = LiPD_to_df (aLiPD)**
# * Convert a LiPD container to 2 pandas dataframes for Data and Metadata, respectively
# - **df_plot (dfD, xCol, [yCols], width=600, height=600)**
# * Convert a data pandas dataframe to an interactive plot using xCol and yCols, respectively as xaxis and yaxis
# * The function relies on bokeh module
# - **PDS_to_html (PDS_inputfile, xCol, [yCols], HTML_outputfile, title=None, width=600, height=600)**
# * Convert a PDS to an html file with Data as an interactive plot and Metadata as a table
# * The function relies on bokeh module
# ### Convert a PDS to pandas dataframes
# In[1]:
import PDSlib
# In[2]:
dfD, dfM = PDSlib.PDS_to_df('PDS_01.xls')
# In[3]:
dfD
# In[4]:
dfM
# ### Convert the pandas dataframes to a LiPD container
# In[5]:
a_LiPD = PDSlib.df_to_LiPD(dfD, dfM, verbose=True)
import json
print json.dumps(a_LiPD, sort_keys=False, indent=4, separators=(',', ': '))
# This exemple comes from the article [Linked Data JSON (JSON-LD) format](https://www.authorea.com/users/17200/articles/19163/_show_article) but somes keys have been changed from column numbers to parameter names accordingly the note #2.
# ### Convert a LiPD container to pandas dataframes
# In[6]:
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
# In[7]:
dfD
# In[8]:
dfM
# ### Convert pandas dataframes to a PDS
# In[9]:
PDSlib.df_to_PDS(dfD, dfM, 'PDS_02.xls')
# PDS_01.xls is identical to PDS_02.xls proof that conversions are correct.
#
# PDS ===> LiPD ===> PDS
# ### Direct conversion from a PDS to a LiPD container
# In[10]:
a = PDSlib.PDS_to_LiPD('PDS_Uemura.xls', verbose=True)
import json
print json.dumps(a, sort_keys=False, indent=4, separators=(',', ': '))
# ### Direct convertion from a LiPD container to a PDS
# In[11]:
PDSlib.LiPD_to_PDS(a, 'PDS_04.xls')
# ### Convert the data pandas dataframe as an interactive plot with [bokeh]( http://bokeh.pydata.org/en/latest/)
# In[12]:
dfD, dfM = PDSlib.PDS_to_df('PDS_Uemura.xls')
# In[13]:
dfM.sort_values(by='Attribute')
# In[14]:
dfD.columns
# In[15]:
PDSlib.df_plot(dfD, 1, [2,3,4])
# ### Convert a PDS to an html file with Data as an interactive plot and Metadata as a table with bokeh
# In[16]:
PDSlib.PDS_to_html("PDS_Uemura.xls", 1, [2,3,4], "PDS_Uemura.html", width=600, height=400)
# [Open the html file created](PDS_Uemura.html)
# ## Below are some tests to deal with values entered as lists
# ### A conversion test from a BibJSON that describes publication metadata
# In[17]:
a_LiPD = [{
"author": [
{"name" : "Thornalley, D.J.R"},
{"name" : "Elderfield, H."},
{"name" : "McCave, N"}
],
"type" : "article",
"identifier" :
{"type": "doi",
"id": "10.1038/nature07717",
"url": "http://dx.doi.org/10.1038/nature07717"}
,
"pubYear": 2009
}]
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
dfM
# ### A conversion test with a GeoJSON
# In[18]:
a_LiPD = { "type": "FeatureCollection",
"features": [
{ "type": "Feature",
"geometry": {"type": "Point", "coordinates": [102.0, 0.5]},
"properties": {"prop0": "value0"}
},
{ "type": "Feature",
"geometry": {
"type": "LineString",
"coordinates": [
[102.0, 0.0], [103.0, 1.0], [104.0, 0.0], [105.0, 1.0]
]
},
"properties": {
"prop0": "value0",
"prop1": 0.0
}
},
{ "type": "Feature",
"geometry": {
"type": "Polygon",
"coordinates": [
[ [100.0, 0.0], [101.0, 0.0], [101.0, 1.0],
[100.0, 1.0], [100.0, 0.0] ]
]
},
"properties": {
"prop0": "value0",
"prop1": {"this": "that"}
}
}
]
}
dfD, dfM = PDSlib.LiPD_to_df(a_LiPD)
dfM
# These last 2 conversions examples are not bidirectional since list items in LiPD containers are transformed using numbers for each item when PDSlib.LiPD_to_df is used:
# - author[] to author.1, author.2, author.3
# - features[] to features.1, features.2
#
# Should lists be allowed in LiPD ? If so, more work is required to recreate a list from numbered items when using PDSlib.df_to_LiPD
# - author.1, author.2, author.3 to author[]
# - features.1, features.2 to features[]
# In[ ]:
# In[ ]: